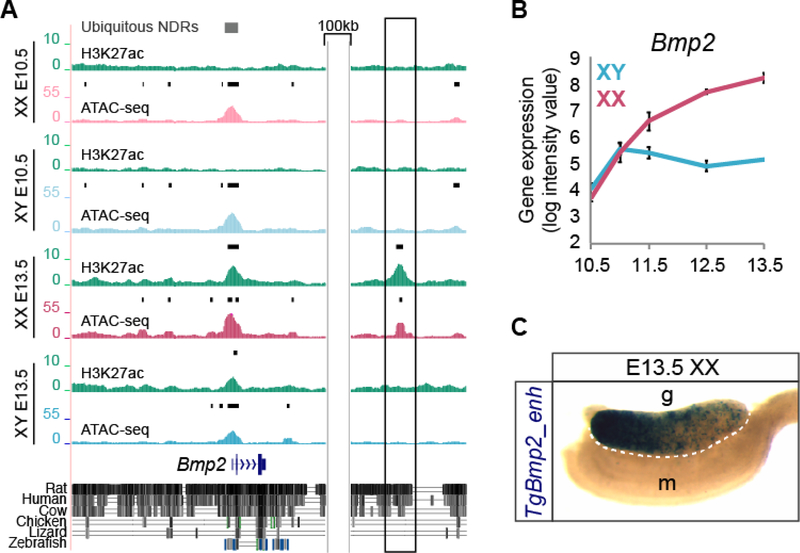
Figure 5. Transient transgenic analysis of a granulosa-specific NDR downstream of Bmp2.
A) Genome Browser tracks of ATAC-seq in XX E10.5 (light pink), XY E10.5 (light blue), granulosa (dark pink) and Sertoli (dark blue) purified cells and corresponding H3K27ac ChIP-seq profiles (green) at the genomic location surrounding Bmp2. Bold black lines above tracks represent significant enrichment compared to flanking regions as determined by HOMER. Grey boxes represent ubiquitous NDRs (top). Sequence conservation between rat, human, cow, chicken, lizard, and zebrafish are shown below. A de novo gonad-specific H3K27ac+ granulosa NDR downstream of Bmp2 (boxed) was selected for transient transgenic analysis and cloned into an hsp68-LacZ reporter cassette. B) Gene expression profile in purified XX (pink) and XY (blue) supporting cells throughout sex determination (E10.5-E13.5) showing that Bmp2 becomes granulosa-specific (Nef et al., 2005). C) An E13.5 XX gonad from a transgenic embryo shows β-galactosidase expression (TgBmp2_enh, dark blue) in the gonad (g), but not the mesonephros (m). TgBmp2_enh expression is stronger in the anterior pole of the gonad (left).