Figure 3: NRL affects cell cycle and controls CCND2 expression.
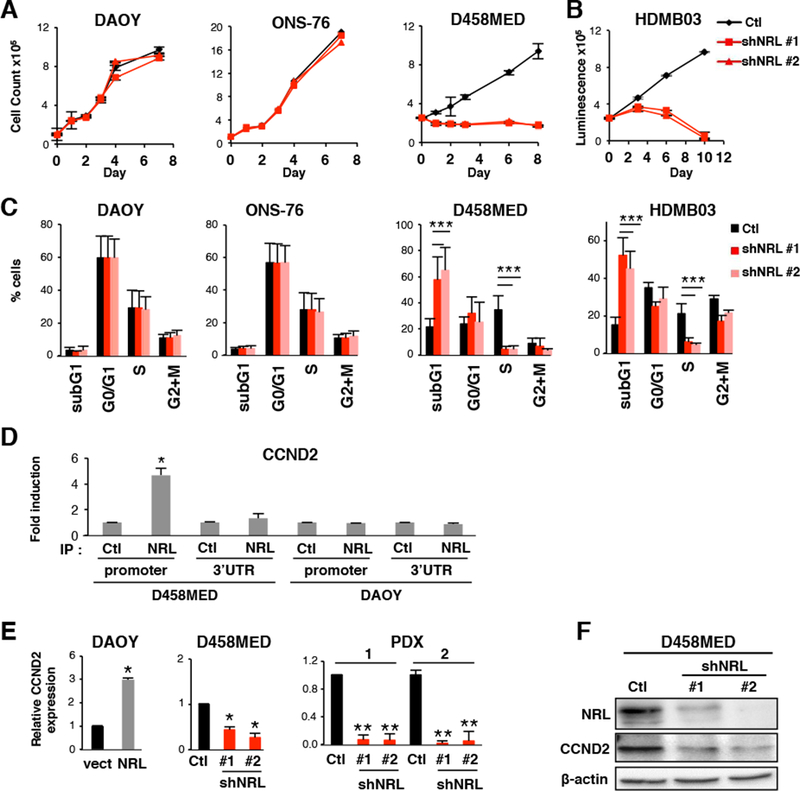
(A-C) Effect of shRNA-mediated NRL knockdown in NRL expressing cells (D458MED, HDMB03 or in non-expressing cells (DAOY and ONS-76). shRNA control (Ctl,black) and two shRNAs targeting NRL were used (shNRL#1 and shNRL#2, red) as indicated. (A) Growth curve analysis and (B) viability assay. (C) Cell cycle analysis by FACS. (D) ChIP analysis was performed by qPCR to analyze the binding of NRL to two different regions of the CCND2 locus, the MARE (Maf-Responsive-Element) containing region of the CCND2 promoter and the 3’ UTR (negative control). NRL antibody (IP NRL) or a control antibody (IP Ctl) were used in D458MED and DAOY cells. (E) Expression level of CCND2 by RT-qPCR in DAOY stably expressing NRL (NRL) relative to control DAOY transfected with the empty vector (vect), in D458MED or in PDX1 and 2 cultures following downregulation of NRL by shRNA (shNRL#1 and #2) relative to control shRNA (Ctl). The level of CCND2 mRNA was set at 1 for controls. (F) Expression level of CCND2 by immunoblot in D458MED expressing NRL-target shRNA (shNRL#1 and #2) and control shRNA (Ctl). Each experiment was performed in triplicate. Bars represent the mean ± SD. The p value was determined by unpaired t test. * p-value < 0.01, **p-value < 0.001, ***p-value<0.0001.
