Abstract
MicroRNAs target mRNAs for cleavage or translational repression. They play a critical role in the progression of malignancies and leukemias including chronic lymphocytic leukemia (CLL). However, microRNA expression levels in Egyptian patients with CLL, and their prognostic value remain elusive. Our main aim was to assess the expression pattern of a panel of microRNAs in CLL patients to create an informative microRNA profile. The study subjects were 40 newly diagnosed CLL patients of both sexes and 40 age and sex matched controls. The expression levels of 12 microRNAs were evaluated by qRT-PCR, including miR-15a, 16, 23b, 24, 29a, 29c, 34a, 146a, 155, 181a, 195, and 221. Flow cytometry was used to determine the expression levels of BCL2, CD38, and ZAP-70 in CLL patients. We identified various degrees of upregulated miRNAs (miR-29a, miR-29c, miR-34a, miR-155, miR-146a, and miR-195) and down-regulated ones (miR-15a, miR-16, miR-23b, miR-24, miR-181a, and miR-221) in CLL patients relative to controls. The mean fluorescence intensity ratio (MFI-R) of BCL2 was recorded and was significantly upregulated in CLL patients compared with normal controls. In addition, inverse correlations were observed between microRNAs (miR-15a, miR-16, miR-155, and miR-195) and BCL2 MFI-R while positive correlations were observed between miR-29a and miR-29c, and BCL2 MFI-R. These findings suggest that these miRNAs regulate BCL2 levels. Moreover, we found that miR-15a, miR-16, miR-155, miR-181a, miR-195 and miR-221 were significantly upregulated, while miR-29a and miR-29c were significantly downregulated in ZAP-70 positive CLL patients. Various miRNAs may play an important role in the pathogenesis of CLL and have the potential to be used for the prognosis of patients with CLL.
Electronic supplementary material
The online version of this article (10.1007/s12288-018-1000-7) contains supplementary material, which is available to authorized users.
Keywords: miRNA, CLL, BCL-2, ZAP-70, CD38
Introduction
Chronic lymphocytic leukemia (CLL) is the most common form of adult leukemia accounting for approximately 30% of all leukemias [1]. Although the first clinical description of CLL was published over 150 years ago, it remains an ‘enigma’ of modern hematology [2].
MicroRNAs (miRNAs) are an abundant class of short regulatory (~ 22 nt) non-coding RNAs that are widely expressed in all eukaryotes, encoded by genes that are presumably transcribed into single or clustered primary transcripts that are processed to produce the mature miRNAs. They are then incorporated into a ribonucleoprotein complex called the RNA-induced silencing complex (RISC) and guide the RISC to the target mRNA. RISC with bound miRNA mediate targeted down-regulation of genes by cleavage of mRNA or by translational repression [3]. Their biological stability, non-invasiveness, and predictive properties make circulating miRNAs ideal biomarkers for cancer.
Human cancer is associated with changes in miRNA expression. It was recently reported that the pattern of miRNA expression varies dramatically across tumor types, and that miRNA profiles reflect the developmental lineage and differentiation state of the tumor [4]. In addition, miRNAs are likely to play critical roles in various aspects of hematopoiesis [5].
Taking the above into consideration, it is no surprise that miRNAs have been found to be instrumental in CLL physiology. In particular, certain miRNAs are known to be downregulated in CLL compared with normal B cells, and distinct miRNA signatures are associated with prognostic factors, disease progression, response to treatment, and various other disease characteristics [6].
In the future, patient-specific therapeutic drugs may be designed for people with CLL harboring abnormalities in miRNA expression in their malignant cells.
Inhibiting overexpressed miRNAs and inducing expression of silenced miRNAs in cancer cells may contribute to selective tumor killing, thus, miRNAs are potential targets for therapy. Loss of miRNA expression in patients with CLL may selectively suppress proapoptotic pathways providing such malignancies with a survival advantage. In fact, some promising results have already been achieved using ‘antagomirs’ in animal models [7].
Determining the involvement of miRNAs in CLL and using this knowledge for patient benefit is challenging but there is no doubt that the time for the “miRNA revolution” in CLL has arrived.
The main aim of the present study was to assess the expression pattern of a panel of miRNAs in Egyptian CLL patients in order to create an informative miRNA profile for CLL and to study their role in pathogenesis as well as their relationship with other prognostic markers and clinical status.
Materials and Methods
Study Design
Our study was conducted with 40 newly diagnosed CLL patients of both sexes that were admitted to Alexandria Main University Hospital. The diagnosis of CLL was established based on current WHO guidelines [8]. Patients were classified according to the modified Rai staging system. Forty normal healthy volunteers of matched age and sex were included in the study as the control group. All participants provided informed consent and the study was approved by the Ethics Committee of the Faculty of Medicine, Alexandria University. Treatment of our patients was based on the International Workshop on Chronic Lymphocytic Leukemia (IWCLL) updated recommendations for the management of CLL in clinical trials and general practice [9].
Our treatment modality was as follows: FCR (Fludarabine, Cyclophosphamide, Rituximab) regimen for 6 cycles in cases indicated and fit for treatment. In case of relapse or progressive disease; patients receive the CHOP regimen. Otherwise, patients with asymptomatic early-stage disease were monitored without therapy until they have evidence of disease progression.
According to the IWCLL consensus response criteria for CLL; complete remission (CR) requires absence of peripheral blood clonal lymphocytes by immunophenotyping, absence of significant lymphadenopathy, absence of hepatomegaly or splenomegaly, satisfactory blood counts and absence of constitutional symptoms. Partial remission (PR) is characterized by a decrease by 50% or more of the pre-treatment lymphocyte count, lymphadenopathy, hepatomegaly, and splenomegaly and adequate blood counts.
Progressive disease during or after therapy is characterized by at least one of the following: progressive lymphadenopathy, denovo appearance or progressive enlargement of hepatomegaly or splenomegaly, An increase in the number of PB lymphocytes by 50% or more. Appearance of any cytopenias, and/or more aggressive transformation. Stable disease patients are those who neither achieved CR or partial response and did not reach progressive criteria. Treatment failure is any outcome other than CR and PR. Relapsed patients are those Patients who after a period of 6 months or more of achieving CR develop disease progression [9].
A full history, thorough clinical examination, and laboratory investigations were carried out on all patients. Complete blood counts (CBC) were performed on an ADVIA-2120i automated cell counter (Siemens Healthcare Diagnostics Inc., USA). Immunophenotyping for patients at diagnosis was performed using a Becton–Dickinson FACS Caliber Flow Cytometer equipped with CellQuest Software (BD Biosciences, San Diego, California, USA). Quantitative determination of miRNA expression was performed by real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR). Follow ups were performed on CLL patients ranging between 7 and 26 months after initial diagnosis until the end of the study with a median of 23.5 months.
miRNA Assay
miRNA analysis was performed using fresh peripheral blood samples collected in K2EDTA tubes. Total RNA, including miRNA isolation was carried out with the miRNeasy Mini Kit (QIAGEN, Maryland, USA, Cat# 217004) according to the manufacturer’s instructions. cDNA was synthesized from the purified RNA samples using the RT2 miRNA First Strand Kit (QIAGEN, Maryland, USA) for microRNA Reverse Transcription according to the manufacturer’s protocol: In a total volume of 10 μL, the reaction mixture was incubated at 37 °C for 2 h and then heated at 95 °C for 5 min on a Techne Touchgene Gradient PCR Thermal Cycler (Techne, Germany). Samples were chilled on ice, and finally 90 μL RNase-free H2O were added and the cDNA was stored at − 20 °C until further processed.
Expression of miRNAs 15a, 16, 23b, 24, 29a, 29c, 34a, 146a, 155, 181a, 195, and 221 was carried out using Custom RT2 profiler miRNA PCR Arrays (Cat. # CAIH0037A QIAGEN sciences, Maryland, USA). A 96-well plate layout was used on a Stratagene Mx3000P QPCR System (Stratagene, Amsterdam, NL, Europe) with RT2 SYBR® Green ROX™ qPCR Mastermix (QIAGEN, Maryland, USA). In a total volume of 425 μL, the reaction mixture was incubated at 95 °C for 10 min for initial denaturation, followed by 40 cycles of PCR, each of which consisted of denaturation at 95 °C for 15 s, annealing at 60 °C for 40 s, and extension at 72 °C for 30 s. The relative expression of each miRNA was calculated with the comparative CT method (2−ΔΔCT) after normalization for the expression of SNORD47 and SNORD48 as endogenous controls.
BCL2, CD38 and ZAP-70 Assay
Flow cytometric analysis was performed on fresh peripheral blood samples collected in K2EDTA tubes to confirm the diagnosis of CLL and for the detection of BCL2, CD38 and ZAP-70 using our CLL panel together with BCL2-FITC (clone 124; Cat# F7053), CD38-FITC (clone T-16; Cat# 07778), and ZAP-70-PE (clone G-4; Cat# 17760) purchased from Dako Cytomation (Denmark). Isotype controls were run with each sample. BCL2 mean fluorescence intensity ratio (MFI-R) was measured for CLL patient B-cells in relation to CD19 negative lymphocytes. A “stain and then lyse/wash” technique was used. Staining was performed according to the manufacturer’s instructions. Briefly, 10 μL of the monoclonal antibodies were added to 100 μL of whole EDTA blood, mixed well, and incubated for 10 min at room temperature. The cells were then washed twice with phosphate buffered saline (PBS), 2 mL lysing solution were added, mixed, and left for 10 min in the dark, followed by two PBS washes. After the last wash, the cells were analyzed. ZAP-70 and BCL2 are intracytoplasmic antigens, and therefore required an extra permeabilization step. The cut-off point of positivity was set to be when > 20% of the gated cells were stained in excess of the background fluorescence in the negative controls.
Statistical Analysis
All statistical analyses were performed using the IBM SPSS software package version 20.0 (Armonk, NY: IBM Corp). Comparison between different groups regarding categorical variables was tested using Chi square test The Kolmogorov–Smirnov, Shapiro and D’agstino tests were used to verify the normality of distribution of variables. For normally distributed data, comparisons between two independent populations were done using independent t test.
Mann–Whitney test was used to compare between two groups for abnormally distributed quantitative variables, while Kruskal–Wallis test was used to compare more than two groups. Spearman coefficient was used to correlate between quantitative variables. Receiver operating characteristic curve (ROC) was plotted for miRNAs, the area under the ROC curve denotes the performance. Multivariate Logistic regression was used to determine the best combination of miRNAs predicting the prognostic status of CLL patients and was used to determine the best combinations of ROC curves. Cox regression analysis was used to evaluate the effect of miRNA expression levels on TTT. Overall survival distributions were plotted using Kaplan–Meier estimates and were compared using the log-rank test.
Principle component analysis (PCA) was used to determine the miRNA with the highest load on the outcome. Significance of the obtained results was judged at the 5% level.
Results
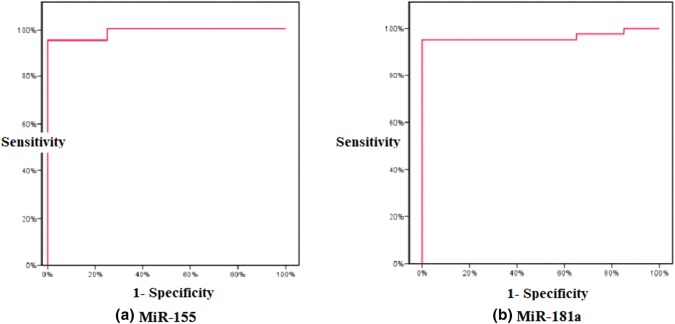
Patients included in the study were 26 males and 14 females, while the control group were 21 males and 19 females. The most common clinical presentation among patients was lymphadenopathy (82.5%) (Table 1), followed by splenomegaly (55.0%) and hepatomegaly (25.0%). Lymphocyte doubling times (LDT) were less than 1 year (< 1Y) in 19 patients (47.5%). From our 40 patients; 28 received treatment according to our treatment modality from which only 10 achieved CR, while the remaining 12 were monitored periodically only for any progress. Comparative marker analysis in CLL samples compared to control samples identified a CLL specific miRNA signature consisting of upregulation of miR-29a, miR-29c, miR-34a, miR-146a, miR-155, and miR-195 and downregulation of miR-15a, miR-16, miR-23b, miR-24, miR-181a, and miR-221 (Supplementary Figure 1). The diagnostic performance of these miRNAs was analyzed with receiver operating characteristic (ROC) curves and the associated area under the curve (AUC). miR-155 (AUC = 0.988), miR-181a (AUC = 0.962) (Fig. 1) and miR-29a (AUC = 0.951) were the best candidate markers to distinguish CLL patients from healthy controls.
Table 1.
Comparison of demographic data, clinical, and laboratory findings between CLL patients and healthy controls
| Patients (n = 40) | Control (n = 40) | Test for sig. | |
|---|---|---|---|
| Sex | |||
| Male | 26 (65.0) | 21 (52.5) | ap = 0.256 |
| Female | 14 (35.0) | 19 (47.5) | |
| Age (years) | 58.40 ± 8.04 | 56.58 ± 8.74 | bp = 0.334 |
| HB g/dL | 11.21 ± 2.70 | 13.03 ± 0.77 | bp < 0.001* |
| WBC (× 103/µL) | 48.70 (10.23–844.80) | 6.73 (4.23–10.0) | cp < 0.001* |
| Platelets (× 103/µL) | 172.50 (12.0–521.0) | 295.0 (176.0–422.0) | cp < 0.001* |
| Absolute lymphocyte count (ALC) (× 103) | 38.484 (13.50–633.60) | 2.244 (1.269–3.50) | cp < 0.001* |
| BCL2 MFI-R | 1.72 ± 0.51 | 0.62 ± 0.14 | bp < 0.001* |
| ZAP-70% | |||
| ZAP-70− (< 20%) | 27 (67.5%) | 40 (100%) | ap < 0.001* |
| ZAP-70+ (≥ 20%) | 13 (32.5%) | 0 (0%) | |
| CD38% | ap < 0.001* | ||
| CD38− (< 20%) | 26 (65%) | 40 (100%) | |
| CD38+ (≥ 20%) | 14 (35%) | 0 (0%) | |
| Lymph nodes | |||
| Axillary and cervical | 14 (42.4%) | ||
| Cervical | 9 (27.3%) | ||
| Generalized | 7 (21.2%) | ||
| Inguinal | 2 (6.1%) | ||
| Paraaortic | 1 (3.0%) | ||
| Modified Rai stage | |||
| Low | 18 (45.0%) | ||
| Intermediate | 10 (25.0%) | ||
| High | 12 (30.0%) | ||
| Time to treatment (TTT) (months) | 6.5 (0.0–26.0) | ||
Normally distributed data was expressed as mean ± SD, while abnormally distributed data was expressed as medians (min–max)
*Statistically significant at p ≤ 0.05
ap: p value for Chi square test; bp: p value for Student’s t-test; cp: p value for Mann–Whitney test
Fig. 1.
ROC curve analysis for a miR-155 and b miR-181a expression levels for discriminating CLL patients (40 cases) from control (40)
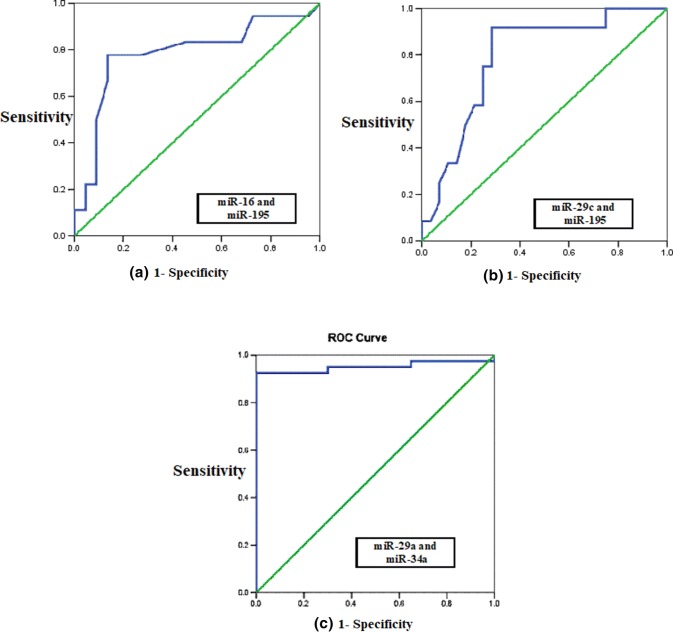
We used logistic regression to determine the best combination of miRNAs to use to distinguish CLL patients from controls. This was achieved by miR-29a and miR-34a that provided a combined AUC of 0.995 (Fig. 2).
Fig. 2.
Combined ROC curve analysis for a miR-16 and miR-195 for predicting the prognostic state (AUC = 0.793), b miR-29c and miR-195 for predicting the treatment requirement (AUC = 0.801), c miR-29a and miR-34a for discriminating CLL patients from the control (AUC = 0.995)
There were no significant differences in miRNA expression levels with regard to age and sex. miR-23b and miR-221 (p = 0.028 and p = 0.034 respectively) were overexpressed in CLL patients with lymphadenopathy (Supplementary Table 1, Supplementary Figure 2)
With regards to hepatosplenomegaly; miR-15a, miR-23b, miR-181a, and miR-195 were overexpressed in patients with hepatomegaly or splenomegaly; miR-16, miR-155, and miR-221 were overexpressed in patients with splenomegaly; miR-34a was overexpressed in patients with hepatomegaly; while miR-29a and miR-29c were underexpressed in patients with splenomegaly or hepatomegaly compared to those patients without organomegaly (Supplementary Table 1, Supplementary Figure 3).
The CLL patients were divided into three stages according to the modified Rai staging system (Table 1). Statistical analysis was performed using the Kruskal–Wallis test to compare miRNA expression from different stages. This analysis showed that a unique signature composed of 11 miRNAs, including miR-15a (p = 0.005), miR-16 (p = 0.006), miR-23b (p = 0.001), miR-24 (p = 0.011), miR-29a (p = 0.001), miR-29c (p < 0.001), miR-146a (p = 0.014), miR-155 (p < 0.001), miR-181a (p = 0.003), miR-195 (p < 0.001) and miR-221 (p = 0.019) had significantly different expression levels among the three stages (Supplementary Figure 4).
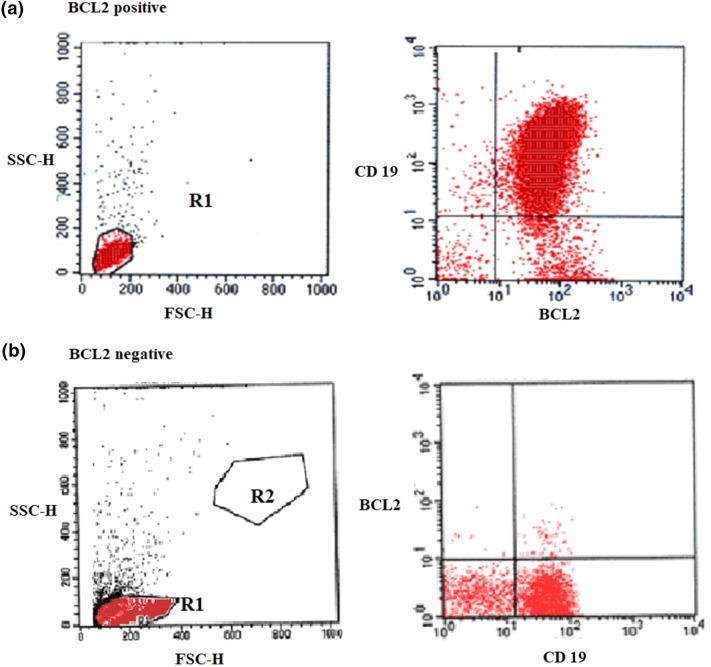
BCL2 MFI-R was recorded for B-cells and CD19 negative non-B lymphoid cells in all participants (Table 1, Fig. 3). Values were significantly higher in CLL patients than in the control group (p < 0.001). BCL2 MFI-R showed statistically significant negative correlations with CD38% (r = − 0.553, p < 0.001) and ZAP-70% (r = − 0.516, p = 0.001) in CLL patients.
Fig. 3.
Flow cytometry dot-plot for BCL2 expression in CLL: a the majority of cells show double positivity for BCL2/CD19 in the upper right quadrant (BCL2 positive). b The cells are CD19+/BCL2—(negative for BCL2)
Correlation between BCL2 MFI-R and miRNAs revealed that miR-15a (p < 0.001), miR-16 (p < 0.001), miR-23b (p < 0.001), miR-24(p = 0.001), miR-155 (p = 0.001), and miR-195 (p = 0.002) showed a statistically significant negative correlation with BCL2 MFI-R in CLL patients. On the contrary miR-29a (p < 0.001) and miR-29c (p < 0.001) showed a statistically significant positive correlation with BCL2 MFI-R in CLL patients (Supplementary Figure 5).
Flow cytometry was used to measure the levels of CD38 and ZAP-70 in CLL cells isolated from 40 patients. Using 20% as the cutoff for defining ZAP-70 and CD38 positivity, the 40 CLL patients were divided into four groups: group 1 (CD38−/ZAP−) included 22 patients, group 2 (CD38+/ZAP−) included 5 patients, group 3 (CD38−/ZAP+) included 4 patients, and group 4 (CD38+/ZAP+) included 9 patients. Groups 2, 3, and 4 were considered to have poor prognoses while group 1 was considered to have good prognoses. The comparison between groups revealed that miR-15a, miR-16, miR-23b, miR-24, miR-146a, miR-155, miR-181a, miR-195, and miR-221 were significantly overexpressed in CLL patients with poor prognosis compared to those with good prognosis. On the contrary, miR-29a, miR-29c and miR-34a were significantly underexpressed in CLL patients with poor prognosis compared to the good prognostic group (Supplementary Table 2).
When the 12 miRNAs were examined using standard principal component analysis (PCA), 5 of them were found to be powerful predictors of CLL prognosis as they achieved a loading exceeding 0.5. The most powerful predictive miRNAs were miR-181a (0.742) followed by miR-15a (0.677), miR-16, miR-34a and lastly; miR-155(0.541).
We tested the possibility of improving miRNA prognostic performance by combining more than one of them. Logistic regression was used to determine the best combination of miRNAs predicting the prognostic status of CLL patients. The best combination was found to be miR-16 and miR-195 (ROC curve AUC = 0.793) (Fig. 2).
miR-15a, miR-16, miR-23b, miR-24, miR-146a, miR-155, miR-181a, miR-195, and miR-221 were significantly overexpressed, while miR-29a and miR-29c were significantly underexpressed in CLL patients with LDT < 1 year compared with those with LDT > 1 year (p < 0.05) (Supplementary Figure 6).
Comparison of CLL patients according to treatment requirement revealed significant overexpression of miR-23b, miR-146a, miR-181a, and miR-221, and significant underexpression of miR-29a, miR-29c and miR-34a in CLL patients who required treatment (Supplementary Figure 7).
Logistic regression was used to determine the best combination of miRNAs that predicted treatment requirement in CLL patients. The best combination was found to be miR-155 and miR-29c (ROC curve AUC = 0.801) (Fig. 2).
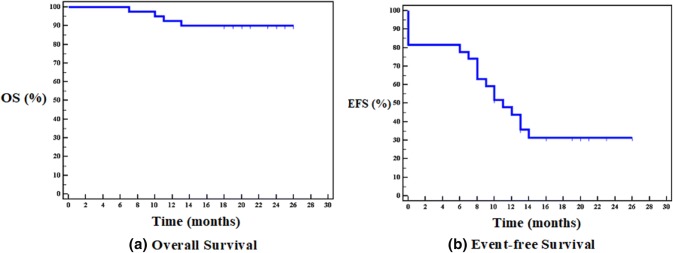
Four of our patients died one from sepsis and the other three died because of refractory/progressive disease (Fig. 4). When miRNA expression levels were compared with regards to CLL survivability, both miR-29c (p = 0.017) and miR-34a (p = 0.003) were found to be significantly lower in CLL patients that had died compared to patients that had survived until the end of the study.
Fig. 4.
Kaplan-Meier curves for a overall survival (40 patients), b event-free survival (28 patients)
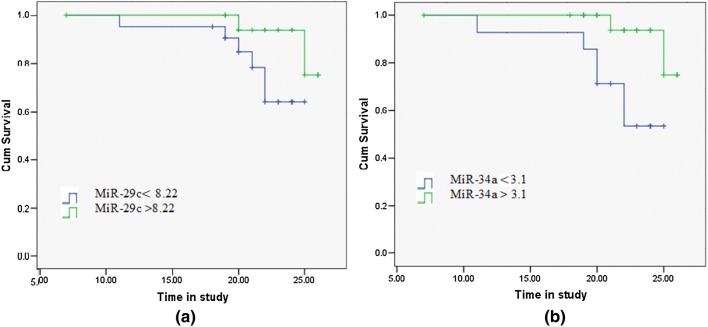
Significant differences in the death rate were found between CLL patients with miR-29c expression levels ≤ 8.22 and > 8.22; (p = 0.05), as well as between miR-34a expression levels ≤ 3.1 and > 3.1; (p = 0.012) over the time of the study. Statistical differences between Kaplan–Meier curves were calculated using the Log-rank test (Figure 5).
To sum up; # A positive relation was found between the bad prognostic markers ZAP-70 and CD38 and the following miRNAs; miR-15a, 16, 23b, 24, 146a, 155, 181a, 195, and 221 that were over expressed in positive cases.
# MiR-23b, 146, 181a, and 221 were over expressed in patients requiring treatment; thus these miRNAs can be used as predictors of treatment.
# miR-155 was found to be significantly affecting TTT by Cox regression analysis, as the model showed that the increase in miR-155 by 1 unit was associated with the increased risk of shorter TTT by about 35%.
# On comparing between patients who achieved CR and the non-responders; miR-29a & miR-34a were significantly under expressed in the non-responders
# Again, miR-29a & miR-34a showed up when we studied the effect on survival as both miRNAs were significantly under expressed in our dead cases.
So, the under expression of both miRNAs can be a predictor of failure of response to treatment and decreased survival.
# Disease progression was found in patients significantly over expressing miR-16, 155, 181a, and 195.
Fig. 5.
Kaplan–Meier curves of CLL patients (40 cases) according to a miR-29c expression levels with a cut-off of 8.22, and b miR-34a expression levels with a cut-off of 3.1
From the above; the use of this panel for each patient can help assessing, TTT, response to therapy, disease progression and survival, and thus treatment can be tailored and modified according to each patient from the time of diagnosis.
Discussion
Extensive testing of all previously published miRNA signatures was not the goal of this study. Rather, we selected a group of 12 miRNAs that were found to be important for CLL pathogenesis in at least 3 independent publications [10–12].
In the present study, the uniformity of miRNA expression changes in CLL patients relative to controls was striking. Though the directionality of changes in expression was consistent for these particular miRNAs, the magnitude of changes varied considerably. Notably, 6 of 12 miRNAs were underexpressed in CLL patients compared to controls, while the other 6 were overexpressed.
In agreement with our results, a distinctive expression profile was found in CLL samples in a study by Calin et al. [13], in which miRNA specific microarrays were used to compare miRNA expression in CLL cells versus normal blood mononuclear cells. Similar results were obtained by Zhu et al. [14], and Zanette et al. [15].
Our results were partially supported by Pallasch et al. [16], who identified 7 upregulated and 19 downregulated miRNAs between CLL patients and healthy donor B cells. The authors identified the upregulation of miR-155, downregulation of miR-181a/b, and heterogeneous upregulation of miR-34a, but did not identify significant downregulation of miR-15 or miR-16. The differences between our results and these may be due to a variably distributed study cohort: the majority of the patients in the study by Pallasch et al. showed a favorable risk profile as assessed by clinical and molecular features, while in our cohort, 45% of patients had an unfavorable risk profile. Differences can also be attributed to the different methodological approaches used (microarray vs. qRT-PCR).
BCL-2 is a central player in the genetic program of eukaryotic cells that promotes survival by inhibiting cell death. We found a significant increase in BCL2 MFI-R in CLL patients versus the control group.
Similar results were reported by McCarthy et al. [17] who confirmed the elevation of BCL2 within CLL cells at the RNA and protein level, and by a number of independent research groups who determined that overexpression of the anti-apoptotic protein BCL2 is a hallmark of CLL [18].
On correlating BCL2 MFI-R with miRNAs expression levels, we found significant negative correlations with miR-15a and miR-16 in our CLL patients. These results are congruent with results from studies by Cimmino et al. [19] and Zhu [20].
Furthermore, we found an inverse correlation between miR-155 and miR-195, and a positive correlation between miR-29a and miR-29c with BCL-2 MFI-R. We did not find any significant correlation between miR-181 and BCL-2, which is not in line with the observations of Neilson et al. [21], In his study, transfection of miR-181a significantly downregulated reporters fused to the 3′ UTRs of BCL-2, suggesting that additional miRNAs are candidate regulators of BCL-2 expression and apoptosis in CLL. Our results are also contrary to those of Cimmino et al. [19], who found an inverse correlation between miR-181a/b and BCL-2 mRNA.
Several reports have suggested that the miRNAs identified in our signature have prognostic value for CLL. Our findings are supported by the observations of Calin et al. [22], who used a miRNA microarray composed of 190 human genes. Similar results were also reported by several other groups of researchers [6, 13, 23].
Importantly, we demonstrated for the first time that miRNA expression levels can individually predict treatment requirement in CLL patients. However, the choice of the cutoff level remains a controversial question. Some investigators determine cutoffs simply by what is found in the literature, but in this study we have opted to use ROC curve analysis to determine the threshold. To our knowledge, we are the first to propose an miRNA expression threshold to predict treatment requirement in CLL patients. At cutoff values of miR-29a (≤ 10.34), miR-29c (≤ 8.22), miR-146a (> 2.19), miR-181a (> 0.1), miR-23b (> 0.15), miR-221(> 0.09), and miR-34a (≤ 3.1), miRNAs achieved variable degrees of sensitivity ranging from 78.57% (miR-181a, 29a) to 50% (miR-34a). The best predictive combination was found to be miR-155 with miR-29c, as the AUC of their combined ROC curve was 0.801. Specificity values ranged from 66.67% (miR-181a) to 100% (miR-34a). Thus, this predictive analysis is likely not yet optimized and these cutoff points should be validated in larger patient cohorts.
Several target genes of miR-181a were found to have oncogenic role like BCL2. Aberrant expression of miR-181a has been associated with many different cancers including MM, AML, breast cancer and glioblastoma [24–27]
MiR-34a, is frequently lost or rearranged in many cancers, including neuroblastoma, CML, AML, and other hematologic malignancies [28]. Members of the miR-34 family are direct p53 targets. MiR-34 genes are up-regulated by p53 and their overexpression in turn causes senescence, apoptosis, or cell cycle arrest by regulating proteins such as BCL2, Cyclin D1, Cyclin E2, cyclin dependant kinase 4 (CDK4), and c-MYC depending on the cell type [10].
In the present study, time from diagnosis until initial treatment (TTT) was calculated. Twenty-eight patients received treatment and their miRNAs expression levels were analyzed by Cox regression analysis to evaluate the effect of miRNA expression levels on TTT. Of the miRNAs studied, only one was significant in this model: miR-155. The model showed that an increase in miR-155 by 1 was associated with an ~ 35% increased risk of a shorter TTT, with all other factors constant. The significance of miR-155 expression on TTT was further confirmed by Kaplan–Meier curve.
MiR-155 represents a typical multifunctional miRNA because it has been implicated in numerous biological functions and disease states including inflammation, cardiovascular disease, adaptive and innate immunity, and neoplastic disease. MiR-155 targets several genes the most important are; the CCAAT/enhancer-binding protein β, Transcription factor 12, Interleukin-17 receptor B oncogenes [29]. MiR-155 overexpression has been described in solid tumors, including lung, colon and breast cancer. Profiling studies demonstrate increased miR-155 expression in both acute myeloid leukemia and CLL.
We also studied the relation between miRNA expression levels and response to therapy through achievement of complete remission (CR) in CLL patients. We found that underexpression of miR-29a and miR-34a was associated with a failure to achieve CR. Similar results were reported by Moussay et al [30] and Zenz et al. [31].
Several miR-29 targets are oncogenes or anti-apoptotic genes MCL-1, TCL1, ten-eleven translocation-1 (TET1), and cyclin dependent kinase 6 (CDK6) genes [32]. The downregulated expression of different miR-29 family members has been reported in multiple malignancies including lung cancer, breast cancer, MDS, and in AML [28].
Stamatopoulos et al. [33] also examined the role of two specific miRNAs (miR-29c and miR-223) in CLL. They examined the expression of these miRNAs in a cohort of 110 CLL patients with CD19+ CD5+ CD23+ phenotypes and determined that downregulation of both miR-29c and miR-223 were associated with poor prognosis, poor treatment-free survival, and overall survival.
Although we followed our cases for up to 26 months, still this is considered a short time interval to adequately judge the prognosis and outcome of a chronic illness as CLL; thus a much longer follow up period is recommended in future studies to better study and assess the prognostic impact of the studied miRNAs
In summary, our data supports previous findings and confirms that miRNAs are involved in the pathogenesis of Egyptian CLL patients. Because the number of cases evaluated thus far is still relatively limited, and no cytogenetic data are available; further analyses using larger prospective patient cohorts together with correlating all data with the cytogenetic studies done to the cases will be required to conclusively define the prognostic relevance of these miRNAs in patients with CLL.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Comparison between cases and control groups regarding miRNAs expression levels (TIFF 52 kb)
Relation between Lymphadenopathy in CLL patients and A) miR-23b expression, B) miR-221 expression (TIFF 25 kb)
Relation between Splenomegaly and miRNAs expression in CLL patients (TIFF 98 kb)
Box plot showing Relation between modified Rai stages and miRNAs expression levels in CLL patients (TIFF 134 kb)
Correlations between miRNAs expression levels and BCL2 MFI-R (TIFF 132 kb)
Relation between LDT and miRNAs expression levels in CLL patients (TIFF 54 kb)
Relation between treatment requirements and miRNAs expression levels in CLL patients (TIFF 53 kb)
Acknowledgements
The authors would like to thank all participating technicians, and our patients, who without them this work could not have been accomplished.
Conflict of interest
The authors declare that they have no conflict of interests.
Ethical Approval
All procedures performed in our study were in accordance with the ethical standards of our institution and national and with the 1975 Helsinki declaration as revised in 2008.
Contributor Information
Nahla Mohamed Gamal Farahat, Phone: 002-01227380886, Email: nahlafarahat@hotmail.com.
Dalal Mohamed Nasr El Din Elkaffash, Phone: 002-01223926483, Email: dalalelkaffash@hotmail.com.
Ashraf Hussein Alghandour, Phone: 002-01223153913, Email: A.alghandour@alexmed.edu.eg.
Rania Shafik Swelem, Phone: 0020-01221768210, Email: raniaswelem@gmail.com, Email: rswelem@yahoo.com, Email: rania.swelem1@alexmed.edu.eg.
Reham Abdel Haleem Abo El-Wafa, Phone: 002-01002413832, Email: rehamhalem@hotmail.com.
References
- 1.Cheson BD, Bennett JM, Grever M, et al. National Cancer Institute-sponsored Working Group guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood. 1996;87:4990–4997. [PubMed] [Google Scholar]
- 2.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 3.Novina CD, Sharp PA. The RNAi revolution. Nature. 2004;430:161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- 4.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 5.Chen CZ, Lodish HF. MicroRNAs as regulators of mammalian hematopoiesis. Semin Immunol. 2005;17:155–165. doi: 10.1016/j.smim.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Papakonstantinou N, Ntoufa S, Chartomatsidou E, et al. Differential microRNA profiles and their functional implications in different immunogenetic subsets of chronic lymphocytic leukemia. Mol Med. 2013;19:115–123. doi: 10.2119/molmed.2013.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang M, Mattes J. Discovery, biology and therapeutic potential of RNA interference, microRNA and antagomirs. Pharmacol Ther. 2008;117(1):94–104. doi: 10.1016/j.pharmthera.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Müller-Hermelink H, Montserrat E, Catovsky D, et al. Chronic lymphocytic leukaemia/small lymphocytic lymphoma. In: Jaffe ES, Harris NL, Stein H, Vardiman JW, et al., editors. World Health Organization classification of tumours: pathology and genetics of tumours of haematopoietic and lymphoid tissues. Lyon: IARC; 2008. pp. 180–182. [Google Scholar]
- 9.Hallek M, Cheson BD, Catovsky D, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on chronic lymphocytic leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111:5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward BP, Tsongalis GJ, Kaur P. MicroRNAs in chronic lymphocytic leukemia. Exp Mol Pathol. 2011;90(2):173–178. doi: 10.1016/j.yexmp.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Nicoloso MS, Kipps TJ, Croce CM, et al. MicroRNAs in the pathogeny of chronic lymphocytic leukaemia. Br J Haematol. 2007;139(5):709–716. doi: 10.1111/j.1365-2141.2007.06868.x. [DOI] [PubMed] [Google Scholar]
- 12.Nana-Sinkam SP, Croce CM. MicroRNA in chronic lymphocytic leukemia: transitioning from laboratory-based investigation to clinical application. Cancer Genet Cytogenet. 2010;203(2):127–133. doi: 10.1016/j.cancergencyto.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Calin GA, Liu CG, Sevignani C, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu DX, Zhu W, Fang C, et al. miR-181a/b significantly enhances drug sensitivity in chronic lymphocytic leukemia cells via targeting multiple anti-apoptosis genes. Carcinogenesis. 2012;33(7):1294–1301. doi: 10.1093/carcin/bgs179. [DOI] [PubMed] [Google Scholar]
- 15.Zanette DL, Rivadavia F, Molfetta GA, et al. miRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz J Med Biol Res. 2007;40(11):1435–1440. doi: 10.1590/S0100-879X2007001100003. [DOI] [PubMed] [Google Scholar]
- 16.Pallasch CP. MiRNA deregulation by epigenetic silencing disrupts suppression of the oncogene PLAG1 in chronic lymphocytic leukemia. Blood. 2009;114:3255–3264. doi: 10.1182/blood-2009-06-229898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarthy BA, Boyle E, Wang XP, et al. Surface expression of Bcl-2 in chronic lymphocytic leukemia and other B-cell leukemias and lymphomas without a breakpoint t(14;18) Mol Med. 2008;14(9–10):618–627. doi: 10.2119/2008.00061.McCarthy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pepper C, Hoy T, Bentley P. Elevated Bcl-2/Bax are a consistent feature of apoptosis resistance in B-cell chronic lymphocytic leukaemia and are correlated with in vivo chemoresistance. Leuk Lymphoma. 1998;28(3–4):355–361. doi: 10.3109/10428199809092690. [DOI] [PubMed] [Google Scholar]
- 19.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu D-X. Aberrant microRNA expression in Chinese patients with chronic lymphocytic leukemia. Leuk Res. 2010;4076:1–5. doi: 10.1016/j.leukres.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 21.Neilson JR, Zheng GX, Burge CB, et al. Dynamic regulation of miRNA expression in orderer stages of cellular development. Genes Dev. 2007;21:578–589. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calin GA, Ferracin M, Cimmino A, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 23.Fulci V, Chiaretti S, Goldoni M, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109(11):4944–4951. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- 24.Debernardi S, Skoulakis S, Molloy G, et al. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21(5):912–916. doi: 10.1038/sj.leu.2404605. [DOI] [PubMed] [Google Scholar]
- 25.Pichiorri F, Suh SS, Ladetto M, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci USA. 2008;105(35):12885–12890. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor MA, Sossey-Alaoui K, Thompson CL, et al. TGF-beta upregulates miR-181a expression to promote breast cancer metastasis. J Clin Invest. 2013;123(1):150–163. doi: 10.1172/JCI64946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen G, Zhu W, Shi D, et al. MicroRNA-181a sensitizes human malignant glioma U87MG cells to radiation by targeting Bcl-2. Oncol Rep. 2010;23(4):997–1003. doi: 10.3892/or_00000725. [DOI] [PubMed] [Google Scholar]
- 28.He L, He X, Lowe SW, et al. microRNAs join the p53 network—another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7(11):819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neilsen PM, Noll JE, Mattiske S, et al. Mutant p53 drives invasion in breast tumors through up-regulation of miR-155. Oncogene. 2013;32(24):2992–3000. doi: 10.1038/onc.2012.305. [DOI] [PubMed] [Google Scholar]
- 30.Moussay E, Palissot V, Vallar L, et al. Determination of genes and microRNAs involved in the resistance to fludarabine in vivo in chronic lymphocytic leukemia. Mol Cancer. 2010;9:115. doi: 10.1186/1476-4598-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zenz T, Mohr J, Eldering E, et al. miR-34a as part of the resistance network in chronic lymphocytic leukemia. Blood. 2009;113(16):3801–3808. doi: 10.1182/blood-2008-08-172254. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Zhang X, Li H, et al. The role of miRNA-29 family in cancer. Eur J Cell Biol. 2013;92(3):123–128. doi: 10.1016/j.ejcb.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Stamatopoulos B, Meuleman N, Haibe-Kains B, et al. MicroRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood. 2009;113:5237–5245. doi: 10.1182/blood-2008-11-189407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison between cases and control groups regarding miRNAs expression levels (TIFF 52 kb)
Relation between Lymphadenopathy in CLL patients and A) miR-23b expression, B) miR-221 expression (TIFF 25 kb)
Relation between Splenomegaly and miRNAs expression in CLL patients (TIFF 98 kb)
Box plot showing Relation between modified Rai stages and miRNAs expression levels in CLL patients (TIFF 134 kb)
Correlations between miRNAs expression levels and BCL2 MFI-R (TIFF 132 kb)
Relation between LDT and miRNAs expression levels in CLL patients (TIFF 54 kb)
Relation between treatment requirements and miRNAs expression levels in CLL patients (TIFF 53 kb)