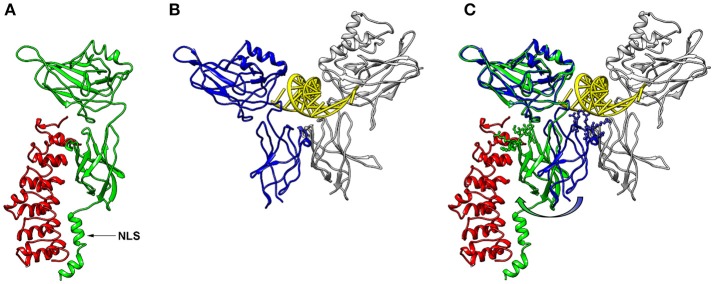
Figure 2.
3D-structures of NF-κB/IκBα and NF-κB/DNA. (A) 3D-Model of a p65-NF-κB fragment (green; amino acids 20–320) bound to IκB (red, amino acids 70–282) generated with Chimera software (20) using the protein database file 1NFI. The position of the nuclear localization sequence (NLS) of p65 is indicated with an arrow. (B) Conformation of a p65 fragment (blue, amino acids 20–291) bound to DNA (yellow) and p50 (gray; amino acids 39–350) forming a characteristic butterfly-like structure (protein database file 1VKX). The p65-fragment, which was crystalized for this structure, lacks the last 29 amino acids of the corresponding structure of (A), but is shown from the same perspective. (C) Superimposed structures of (A, B), illustrating the conformational switch of p65 between the IκB- and the DNA-bound form (green and blue, respectively). The amino acid side chains of the lower p65 wing, which come closer than 0.5 nm to the DNA in the DNA-bound form, are shown in ball-and-stick manner. These side chains are turned away in the IκB-bound form as depicted with an arrow.