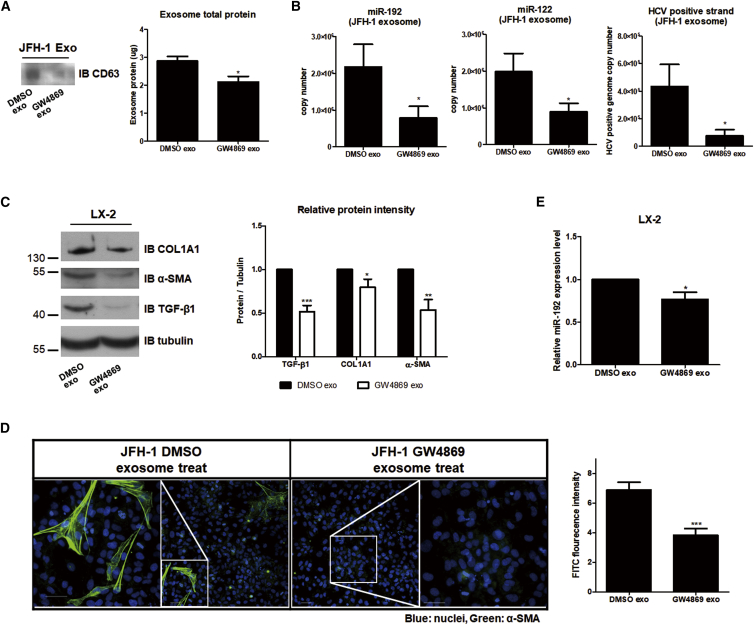
Figure 5.
Effects of Blocking Exosome Release on HSCs
Effects of exosomes from JFH-1 stable cells treated with the exosome release inhibitor GW4869 (10 μM) (GW4869 exo). Control exosomes from untreated JFH-1 stable cells are referred as DMSO exo. (A) Comparison of CD63 (left) or exosome total protein levels using a BCA assay (right) within each exosome is shown. (B) Levels of miR-192 (left: 2.18 ± 1.62 × 106 and 7.82 ± 8.46 × 105 copies per 20 μL of control DMSO exo and GW4869 exo, respectively), miR-122 (middle: 1.98 ± 0.99 × 106 and 8.96 ± 4.68 × 105 copies per 20 μL of DMSO exo and GW4869 exo, respectively), and HCV RNA (right: 4.33 ± 3.21 × 105 and 7.73 ± 8.51 × 104 per 20 μL of DMSO exo and GW4869 exo, respectively) within each type of exosomes were quantified. (C) Immunoblot analysis of the fibrogenic markers COL1A1 and α-SMA, as well as TGF-β1, in LX-2 cells treated with each exosome type (left) is shown. Protein levels were quantified relative to those of tubulin (right). (D) LX-2 cells were treated with each exosome type for 72 h, fixed, stained, and visualized via fluorescence microscopy. Representative images of α-SMA and DAPI staining in each cell are shown (left). Scale bars represent 50 μm. Intensity of α-SMA staining was quantified from more than five fields in three independent experiments (right). Data are also shown in Figure S6. (E) Intracellular miR-192 levels in LX-2 cells treated with each exosome type were quantified. All data are presented as the means of at least three independent experiments, each performed in triplicate. Error bars represent the SEM. p values were determined using a one-tailed unpaired Student’s t test. *p < 0.05; **p < 0.01; ***p < 0.001.