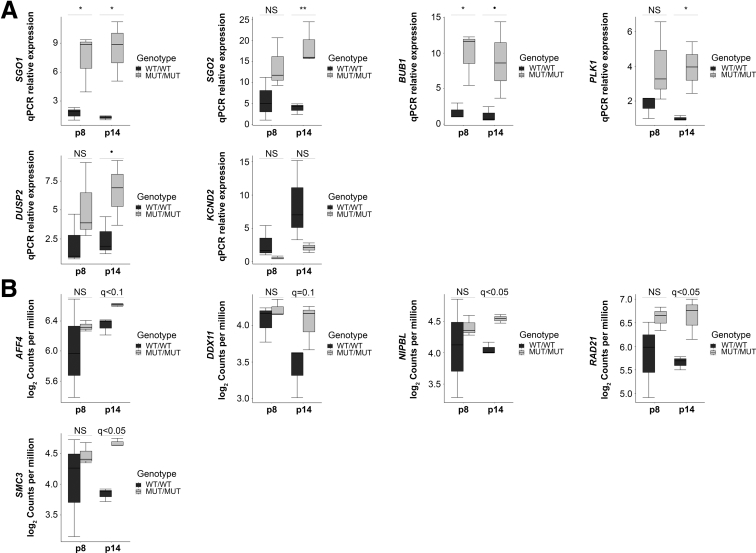
Figure 2.
Candidates validation and expression profile of cohesinopathies genes. (A) Reverse transcription quantitative PCR (qPCR) validations of SGO1, SGO2, BUB1, PLK1, DUSP2, and KCND2 expression at early (p8) and late (p14) passage. Error bars signify SD. For each condition, the experiment was performed on N = 3 independent biological replicates in technical replicates. Significance was calculated using 1-way analysis of variance with the Bonferroni post-test (•P < .1, *P < .05, and **P < .01). (B) mRNA expression levels of genes involved in cohesinopathies at early (p8) and late (p14) passage. AFF4, CHOPS syndrome; DDX11, Warsaw breakage syndrome; NIPBL, RAD21, and SMC3, Cornelia de Lange syndrome. Error bars signify SD. P values were corrected for multiple testing using the Benjamini–Hochberg method (q value). For each group, N = 3 independent biological replicates. MUT, K23E; WT, wild-type.