Table 4.
E3 protein ligand, structure of the linker and POI ligand with a description of the properties research for FKBP12, E3 ligases, an enzyme, a Tau degrader and a recent non-natural fusion protein.
| Table entry | E3 protein (ligand) | Linker | POI ligand | POI | Potency/efficacy | Reference |
|---|---|---|---|---|---|---|
| 1. | CRBN (thalidomide) |
n-butyl and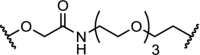
|
 |
FKBP12 | 80% reduction of FKBP12 at 0.1 μM and 50% reduction at 0.01 μM in MV4-11 cells | Winter [6] |
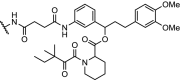
| 2. | VHL (9) | 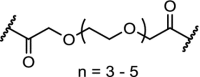 |
(9) | VHL | CM11 (n = 5) induced complete depletion of VHL after 4 h at 10 nM. Potent, long-lasting and selective degradation of VHL, with DC50 of < 100 nM | Maniaci [117] |
| 3. | VHL (9) | 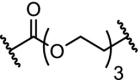 |
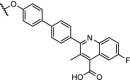 |
DHODH | IC50 for DHODH 93 nM. No degradation observed. Linker optimization needed to target the inner mitochondrial protein. | Madak [121] |
| 4. | Keap1 (Keap1 binding peptide)* | GSGS peptide | YQQYQDATADEQG | Tau | Poly-D-arginine was added for cell penetration. Strong in vitro binding with Keap1 and Tau. Keap1-dependent degradation by enhancing the ubiquitination of Tau. | Lu [124] |
| 5. | CRBN (thalidomide) | 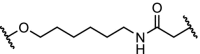 |
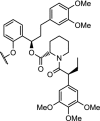 |
FKBP12F36V Fusion proteins | Degradation of a panel of fusion chimeras with FKBP12F36V including: BRD4, HDAC1, EZH2, Myc, PLK1 and KRASG12V. Rapid degradation in vivo was shown | Nabet [33] |
| 6. | CRBN (pomalidomide) | 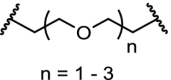 |
pomalidomide | CRBN | The homo-PROTAC with n = 2 was identified as the most potent degrader. Degradation observed at 10 nM after 16 h. Hook-effect observed at 100 μM | Steinebach [118] |
The structure of the CRBN ligands and the VHL ligand (9) are shown in Fig. 2. *Keap1 binding peptide: Ac-LDPETGEYL-OH.
