Figure 1.
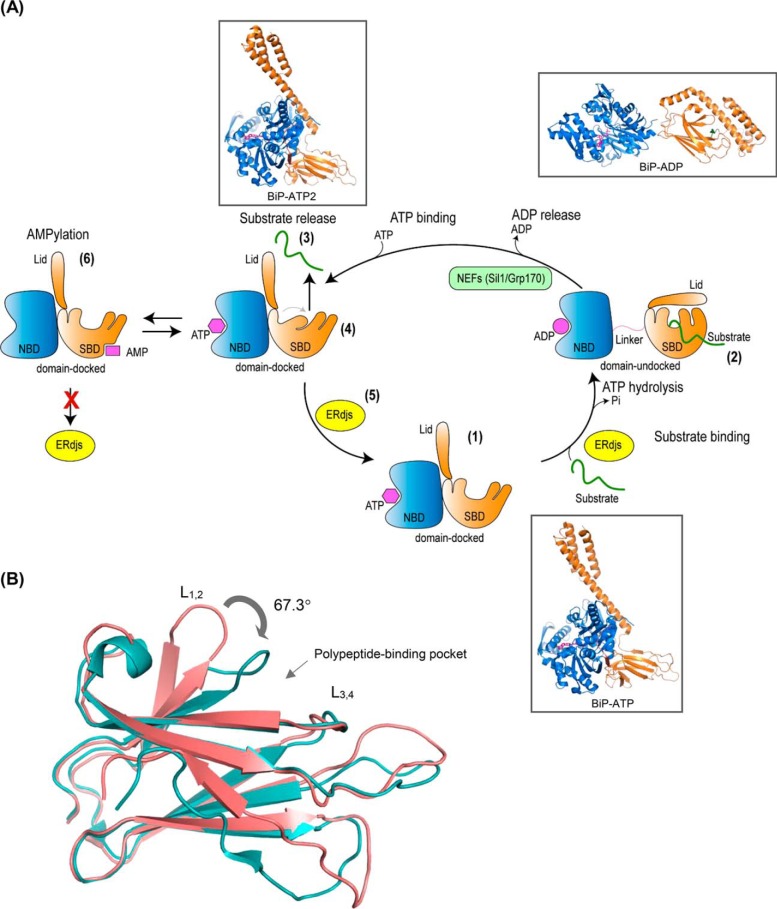
A, BiP ATPase cycle. Step 1, in the ATP-bound form, the nucleotide-binding domain (NBD) (blue) and the substrate-binding domain (SBD) (orange), with its lid open, are docked to each other resulting to a form with high-substrate binding and release kinetics and low-substrate affinity. Step 2, upon ATP hydrolysis, the NBD and SBD become undocked, and the lid of the SBD closes providing a form that has high-substrate affinity but slow binding and release rates. This cycle is regulated by ER-localized DnaJ cofactors (ERdjs) that interact with unfolded proteins and transfer them to the ATP-bound form of BiP, while simultaneously triggering ATP hydrolysis. Step 3, substrate is released with the help of nucleotide-exchange factors (NEFs) that stimulate the release of ADP. Step 4, binding of ATP causes a conformational change in the SBD resulting in a more tightly compacted conformation that is thought to “squeeze” the substrate out. Step 5, interaction with ERdjs reorders the polypeptide-binding pocket of the BiP–ATP2 SBD, readying it to interact with another substrate. Step 6, BiP is post-translationally modified through AMPylation, and this causes the protein to be inactive. AMPylated BiP adopts a “domain-docked” structure similar to that of the ATP-bound state even in the apo- or ADP-bound state and is unable to interact productively with ERdjs. Ribbon representations of the structures (insets): ATP-bound BiP with the polypeptide-binding pocket open (BiP-ATP) (PDB 5e84 (8)); ADP-bound BiP from the structures of the isolated NBD (PDB 5evz (102)); SBD (PDB 5e85 (8)); and ATP-bound BiP with the polypeptide-binding pocket fully closed (BiP-ATP2) (PDB 6asy (8)). B, overlay of the SBD of BiP–ATP and BiP–ATP2. Comparison of SBDβ of BiP–ATP (deep salmon, PDB 5e84) and BiP–ATP2 (teal, PDB 6asy) with their superposition based on Cαs of β3–β7. The peptide-binding loops L1, 2 and L3, 4 are shown. The main difference of the two conformations is that the L1, 2 in BiP–ATP2 is rotated more than 60° compared with that of BiP–ATP resulting in a fully closed polypeptide-binding pocket.