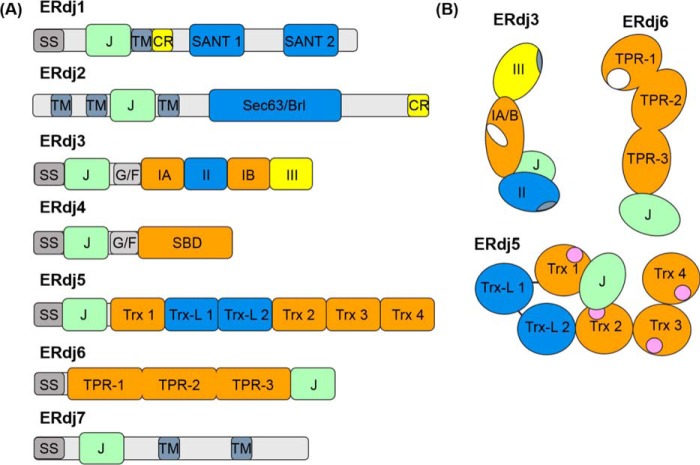
Figure 2.
Domain structure of ERdj family members. A, domain arrangements of primary sequences of each of the known ERdj proteins. Abbreviations used are as follows: SS, signal sequence; J, J domain; TM, transmembrane region; CR, charged amino acid region; SANT, SANT domain; Sec63/Brl, Sec63/Brl domain; G/F, glycine/phenylalanine-rich flexible linker region; IA and IB, bifurcated substrate-binding domain; II, cysteine-rich domain; III, dimerization domain; SBD, substrate-binding domain; Trx, thioredoxin domains; Trx-L, thioredoxin-like, enzymatically inactive domain; TPR, tetratricopeptide repeat domain (each containing three subdomains). B, cartoons representing orientation of the various domains of ERdj5 (72, 73) and ERdj6 (93, 94) based upon known structures. ERdj3 layout was derived from a combination of the solved structure of domain I–III of the yeast homologue Ydj1 (38, 39) and negative strain EM of ERdj3 showing the relative orientation of the J domain (43). The structures of ERdj1, -2, -4, and -7 are unsolved. The following functional regions are also indicated: ERdj3 and ERdj6 substrate-binding sites (white), ERdj3 dimerization/tetramerization regions within domains III and II respectively (blue-gray), and ERdj5's redox-active Trx motifs (pink).