Figure 3.
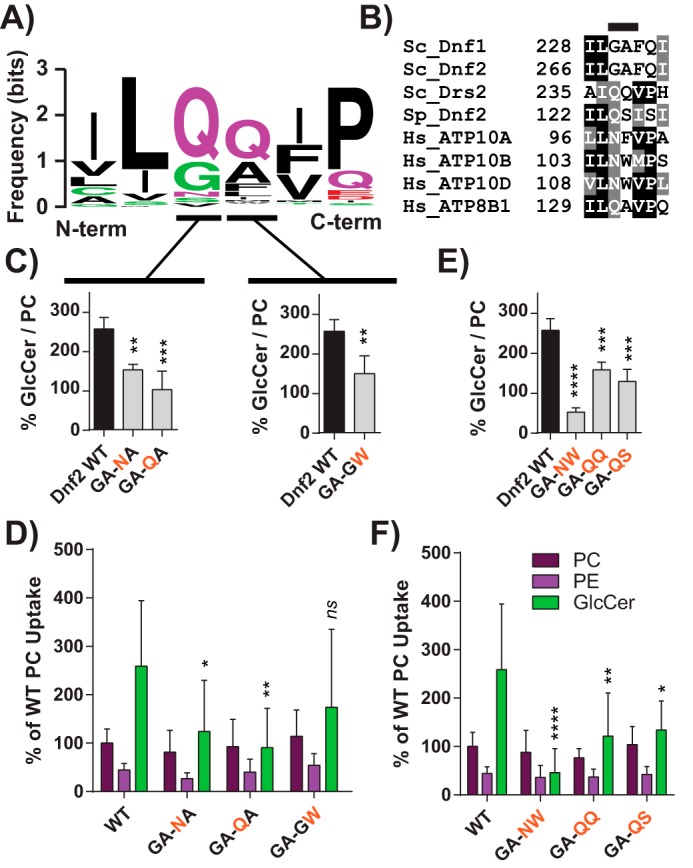
The exofacial TM1 “GA motif” facilitates Dnf2 selection and preference for GlcCer. A, a sequence logo was created from an alignment of TM1 of P4-ATPases from different organisms, with letter size representing residue frequency and color denoting chemical differences. Hydrophilic residues are shown in green and purple, acidic residues in red, and hydrophobic residues in black. B, a focused alignment comparing a region of TM1 from S. cerevisiae, H. sapiens, and S. pombe, highlighting the GA motif. C and D, dissecting the first and second positions of the GA motif reveals that substitutions in both positions can reduce GlcCer preference (C) but do not alter PC or PE recognition (D). E and F, double substitutions were created to examine S. pombe and H. sapiens sequences in the context of the S. cerevisiae Dnf2. These compound mutations reduced GlcCer preference (E) and selection (F) without altering the known glycerophospholipid substrates (F). Variance was assessed among data sets using one-way ANOVAs, and comparisons with WT were made with Tukey's post hoc analysis. Although first- and second-position GA analyses are presented in different panels (C), their statistical variance was tested together. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. Error bars, S.D.
