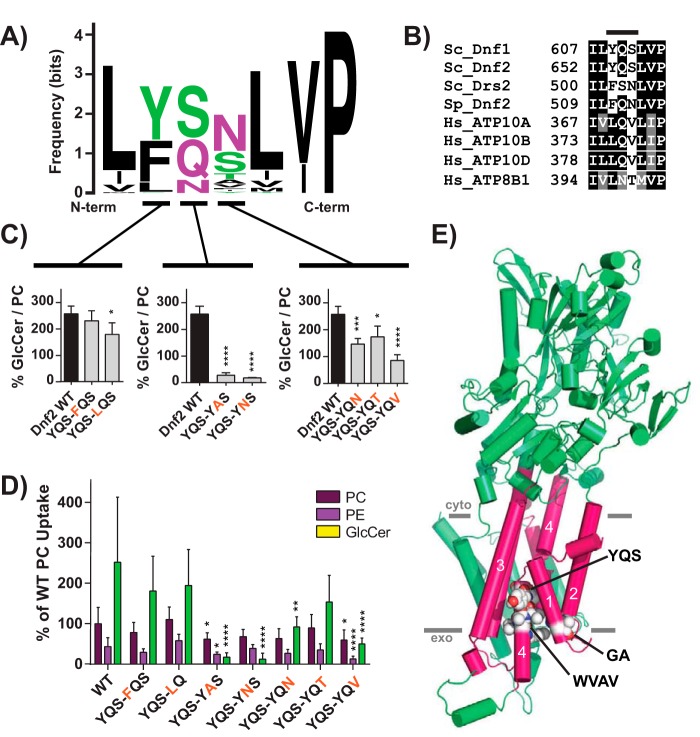
Figure 4.
A Pro−4 glutamine in TM4 of Dnf2 is required for GlcCer transport. A, a sequence logo was created from an alignment of TM4 of P4-ATPases from different organisms, with letter size representing residue frequency and color denoting chemical characteristics. Hydrophilic residues are shown in green and purple, acidic residues in red, and hydrophobic residues in black. B, a focused alignment comparing a region of TM4 from S. cerevisiae, H. sapiens, and S. pombe, highlighting the YQS motif that was previously altered in S. cerevisiae Dnf1 (Fig. S7). C and D, the three YQS positions were tested for their influence on GlcCer preference (C) and selection (D), revealing that the central glutamine was the strongest determinant of GlcCer transport. E, homology model of Dnf1 with TM1–6 shown as pink cylinders and the rest of the protein colored green. GA, YQS, and WVAV motifs are represented by spheres and colored by element. PM boundaries are indicated. Variance was assessed with one-way ANOVAs, and comparisons with WT were made with Tukey's post hoc analysis. Although the YQS positions in C are presented in separate panels, their statistical variance was tested together. n ≥ 9, ± S.D. (error bars). *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.