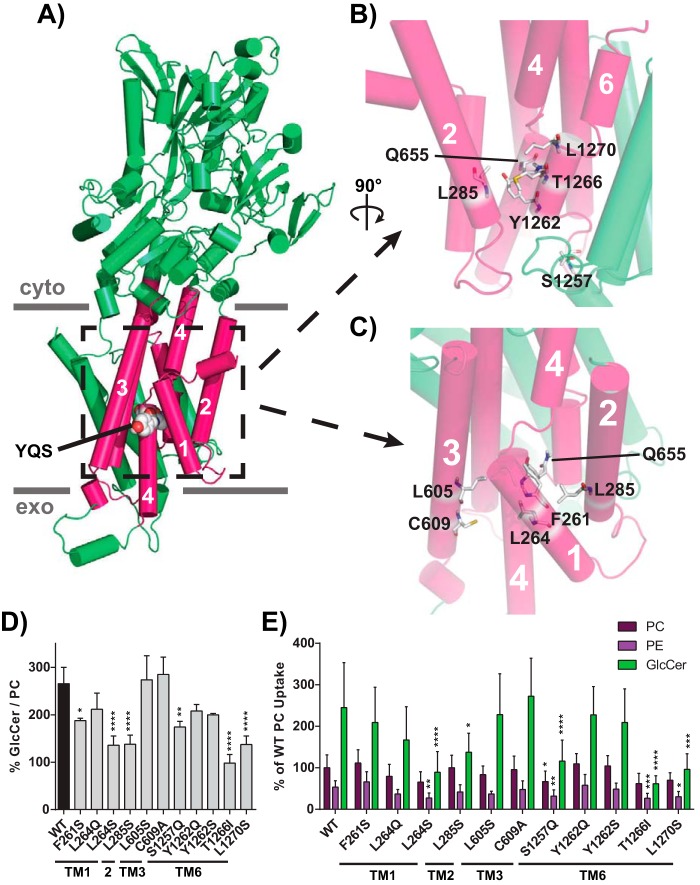
Figure 5.
Residues modeled in proximity to the Pro−4 glutamine influence GlcCer transport. A, homology model of Dnf1 (PDB code 3W5D) with TM1–6 shown as pink cylinders, the rest of the protein colored green, and the YQS motif represented by spheres and colored by element; PM boundaries are indicated. B, a 90° rotated and enhanced view of the peri-Gln655 region formed by TM2, -4, and -6. C, enhanced view of TM1, -2, -3, and -4, which surround Gln655. Residues were selected for mutagenesis by identifying positions that were predicated to be planar with the YQS motif and are shown in sticks and colored by element. D and E, GlcCer preference (D) and selection (E) were examined for all positions, revealing that TM1, TM2, and TM6 positions were capable of altering GlcCer transport. One position required chemical specificity to alter GlcCer transport (Leu264). Variance was assessed among data sets using one-way ANOVAs, and comparisons with WT were made with Tukey's post hoc analysis. n ≥ 9, ± S.D. (error bars). *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.