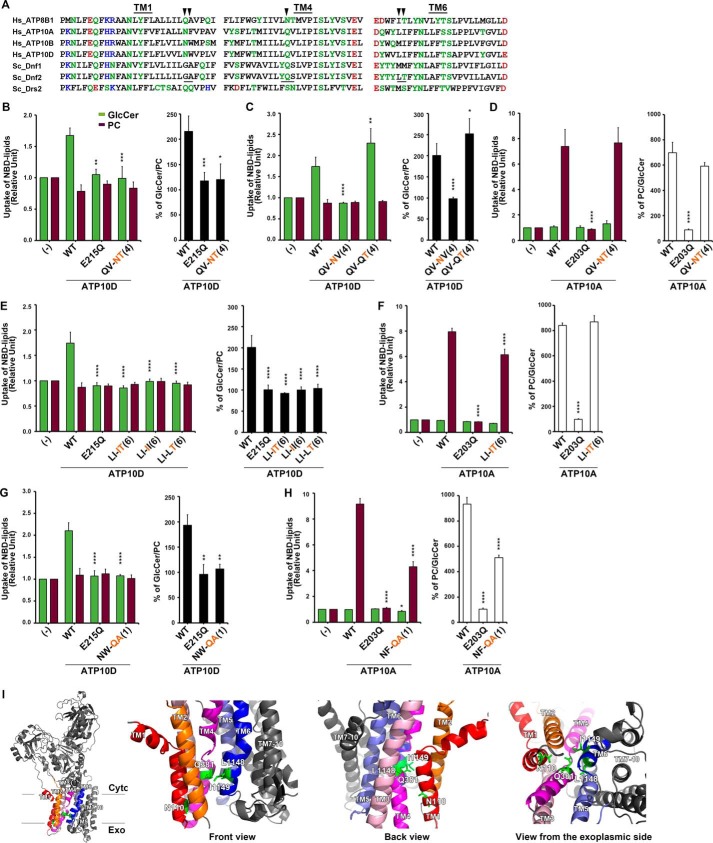
Figure 6.
Structure–function analysis of the H. sapiens ATP10D substrate pathway demonstrates primary structural conservation of TM4 Pro−4 position in GlcCer transport. A, sequence alignments of TM1, TM4, and TM6 of P4-ATPases are shown. Hydrophilic residues are indicated in green, red (negatively charged), and blue (positively charged), and hydrophobic residues are indicated in black. Three motifs, which were required for GlcCer preference in Dnf2, are underlined. The arrowheads indicate amino acids that were critical for ATP10D to transport GlcCer. B–H, NBD-lipid uptake was measured in HeLa cells stably expressing C-terminally HA-tagged ATP10A (WT), ATP10D (WT), each mutant (indicated), and parental cells (−); TM is numbered in parenthesis. B–D, the glutamine at TM4 Pro−4 was critical for ATP10D to transport GlcCer but was dispensable for ATP10A to transport PC. E and F, the motif in TM6 was critical for GlcCer transport of ATP10D but was dispensable for PC transport of ATP10A. The experiments of panels E were performed together with panels C, and thus graphs (−) and WT of E are equivalent to those of C. Graphs display averages from three independent experiments ± S.D. (error bars). -Fold increase of NBD-lipid uptake compared with parental cells (−) is shown. A one-way ANOVA was performed to assess variance in B–H, and comparisons with WT were made with Tukey's post hoc analysis. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. I, TM1, -2, -3, -4, -5, and -6 are indicated in red, orange, pink, magenta, purple, and blue, respectively, and others are indicated in gray. Critical residues for GlcCer transport are indicated by green sticks.