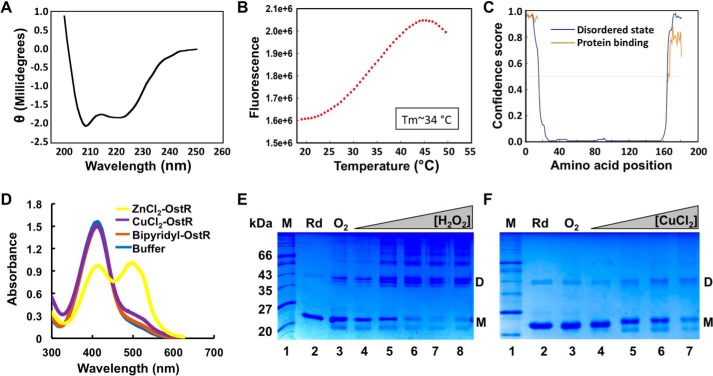
Figure 5.
Characterization of OstR. A, far-UV CD spectrum of OstR. Ellipticity is presented in machine units (millidegrees). B, thermal denaturation of OstR determined by differential scanning fluorometry. Fluorescence intensity reflects binding of SYPRO Orange to hydrophobic regions of denatured protein as a function of temperature. C, prediction of intrinsically disordered regions (blue) and likelihood of disordered regions participating in protein interactions (orange). The y axis reflects confidence score, with values >0.5 considered relevant. D, release of metal ion from denatured OstR determined by absorbance of PAR. Absorbance at 416 nm corresponds to uncomplexed PAR, whereas absorbance at 520 nm reflects formation of PAR–metal ion complex. E and F, OstR oxidation by H2O2 (E) and CuCl2 (F). In both images, the left lanes show protein marker (kDa), lanes 2 show protein incubated with DTT (Rd; species migrating at ∼43 kDa is residual oxidized protein), and lanes 3 show air-oxidized protein (O2). E, lanes 4–8, increasing concentration of H2O2 (10 μm to 2 mm). F, lanes 4–7, increasing concentration of CuCl2 (10 μm to 2 mm).