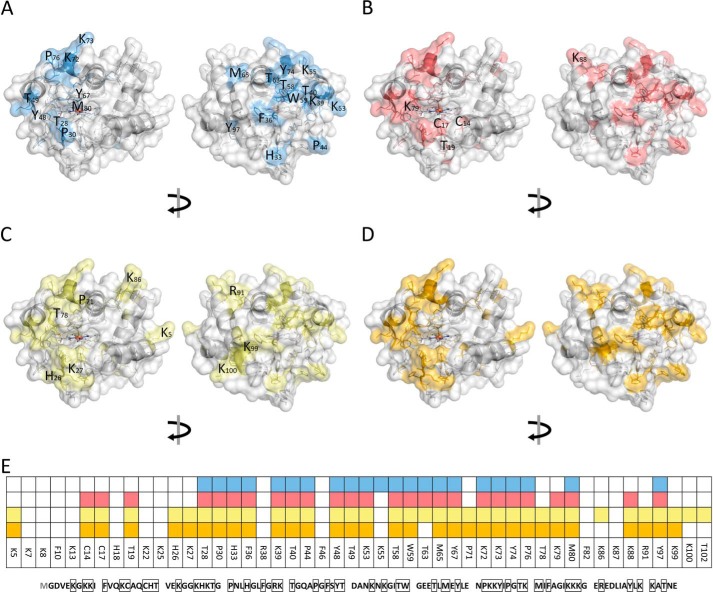
Figure 7.
Modification of cytochrome c identified by LC-MS/MS in control (A), H2O2 (B), TLCL (C), and H2O2 and TLCL (D) incubations and a summary of all (E). Cyt c (100 μm) was incubated in ammonium bicarbonate buffer alone or in the presence of H2O2 (1 mm), TLCL (10 mm), or a combination of both for 30 and 60 min. Proteins were separate by SDS-PAGE, digested with trypsin, and analyzed by LC-MS/MS. PTMs were identified from obtained tandem mass spectra using a search engine–assisted database search. Tertiary structure elements of the protein are shown in cartoon representation, and the surface is shown as semitransparent. Amino acid residues representing possible PTM sites are shown as sticks whereby the residues on which modifications were detected are shown in blue (autoxidation), red (oxidation after incubation with H2O2), green (TLCL), and orange (H2O2 and TLCL). For B, C, and D, only new modifications sites not present in untreated samples are marked with the amino acid one-letter code and position. A model based on Protein Data Bank code 2b4z (cyt c from bovine heart; 1.5-Å resolution) was used and modified with PyMOL version 0.99.