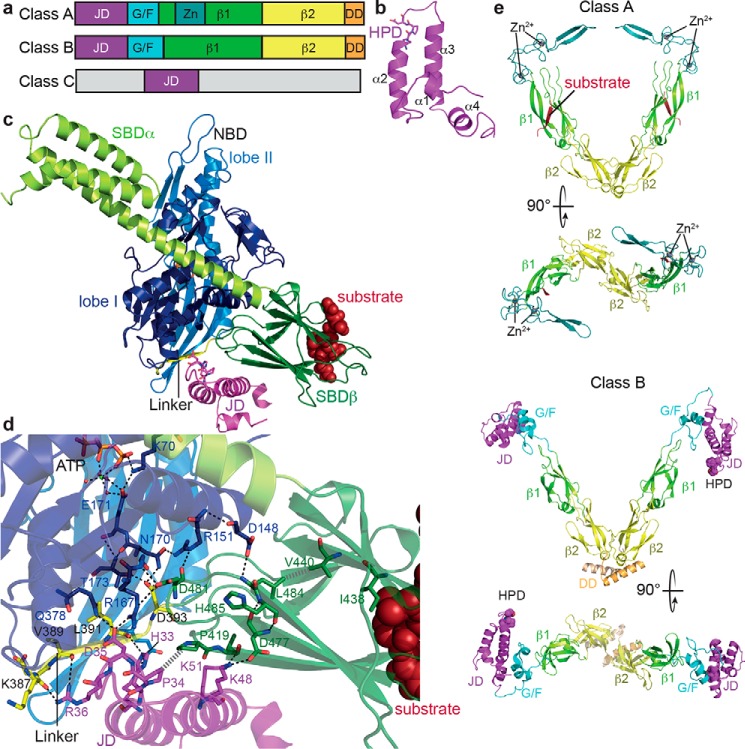
Figure 3.
Structure and function of JDPs. a, domain organization of JDPs; JD, J-domain; G/F, glycine-phenylalanine–rich region; Zn, zinc finger; β1, first β-sandwich domain; β2, second β-sandwich domain; DD, dimerization domain. b, NMR structure of the J-domain of E. coli DnaJ (PDB code 1XBL (122)) indicating the four helices of the helical hairpin and the HPD motif. c, structure of the ATP-bound open conformation of E. coli DnaK in complex with the J-domain of E. coli DnaJ (PDB code 5NRO (82)); NBD lobe I, dark blue; NBD lobe II, light blue; linker, yellow; SBDβ, dark green; SBDα, light green; J-domain, purple. The SBDβ was aligned with the structure of the SBD in complex with a substrate peptide (PDB code 1DKX (10)) to show how peptides (dark red) bind to the ATP-bound open conformation. d, zoom into the structure presented in c showing residues involved in J-domain–DnaK interactions and in substrate and DnaJ-mediated stimulation of the ATPase activity as sticks, polar contacts as black dashed lines, and hydrophobic interactions as gray hatched lines. e, cartoon representation of the β-sandwich domains and the zinc finger of the class A JDP Saccharomyces cerevisiae Ydj1 (PDB code 1NLT (86); dimerization domain is missing in the structure) and the full structure of the class B JDP T. thermophilus DnaJ (PDB code 4J80 (123); J-domain, purple; G/F region, cyan; zinc, dark teal; β1, light green; β2, yellow; dimerization domain, orange.