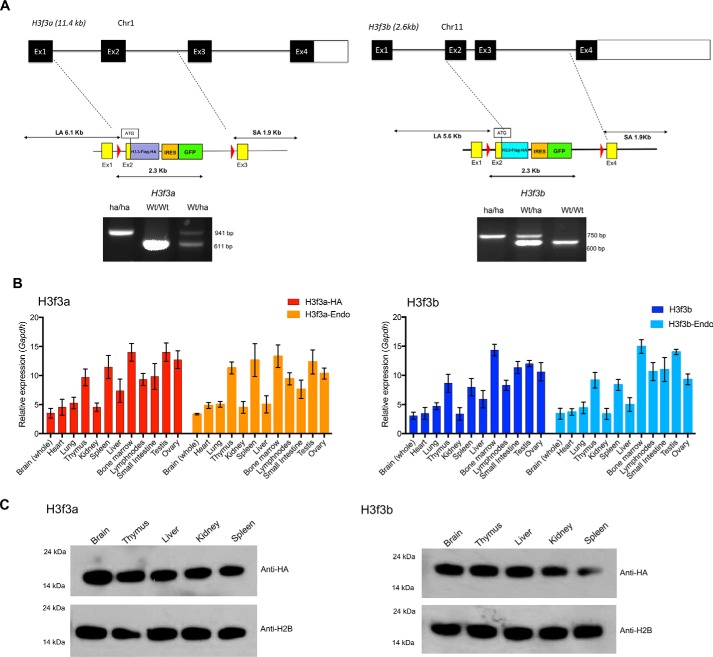
Figure 1.
Knockin mice expressing HA-tagged H3.3. A, top panel, targeting strategy for replacing the endogenous H3f3a and H3f3b loci with the H3.3-HA cDNA cassettes. The exon–intron organization of the endogenous H3f3a and H3f3 loci. Below are the schematic structure of targeting vector and the 5′ and 3′ site arms. The targeting vectors and H3.3 cDNA fused to FLAG-HA, free EGFP, and drug selection markers. Bottom panels, identification homozygous knockin (ha/ha), heterozygous (ha/Wt), and WT (Wt/Wt) pups. Quantitative PCR–based genotyping was performed with tail DNA using primers in Fig. S1C. B, expression of endogenous H3.3 and knockin H3.3-HA mRNA in heterozygous knockin mouse. qRT-PCR detection of mRNA from the H3f3a(ha/wt) (left panel) and H3f3b(ha/wt) loci (right panel) in indicated adult tissues. H3f3a-HA and H3f3b-HA are transcripts from knockin loci, and H3f3a-Endo and H3f3b-Endo are transcripts from endogenous loci. Expression levels were normalized to Gapdh. The values represent the averages of three independent analysis using tissues from 8-week-old mice. C, immunoblot detection of H3.3-HA proteins, from knockin mice. 500 ng of histones prepared from 8-week-old heterozygous mice (H3f3a-HA on the left and H3f3b-HA on the right) were tested by immunoblot using anti-HA or anti-H2B antibody.