Figure 3.
Identification of Potential CFTR-Targeting miRNAs Induced by TGF-β1
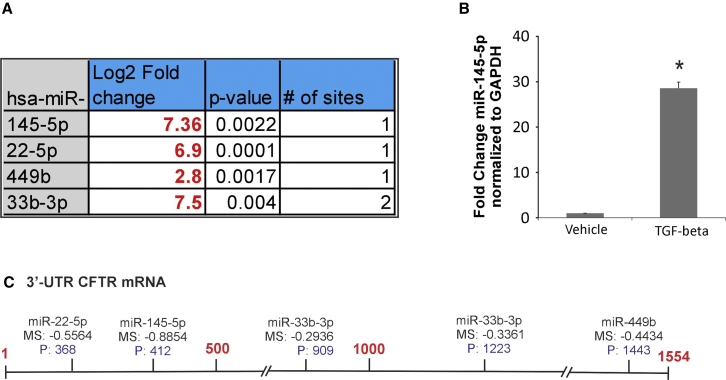
(A) miRNAs targeting CFTR were identified by a combination of target site prediction algorithm miRanda (miRSVR scores <−0.5) and miRTARbase (database of experimentally validated miRNAs). miRNAs are listed in order of their scores starting with the best. (B) Array validation for miR-145-5p. NHBE ALI cultures were treated with TGF-β1 (10 ng/mL) or vehicle (as control). Total RNA was analyzed for miR-145-5p expression using qRT-PCR and normalized to GAPDH. TGF-β1 treatment upregulates miR-145-5p in NHBE ALI cultures. n = 3 different lungs. *Significant (p < 0.05) from control. (C) Schematic of the putative target sites of the identified miRNAs on the 3′ UTR of CFTR mRNA.