Figure 1.
MART-1 Homologs and More Diverse Epitopes Recognized by the DMF5 TCR
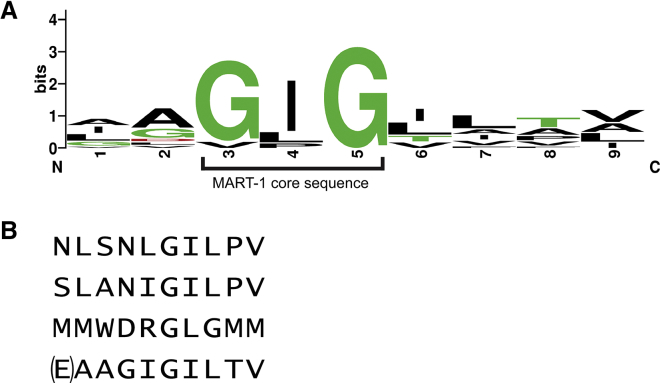
(A) Sequence logo of the previously identified nonameric MART-1 homologs shown in Table 1 that cross-react with MART-1-specific TCRs.36 The GIG sequence spanning positions 3–5, referred to as the MART-1 core sequence, is highlighted. The logo excludes the N-terminally extended MART-1 decamer. The logo was created using WebLogo.76 (B) Divergent epitopes also recognized by DMF5. The top peptide is the NLS epitope, identified via yeast display screening. The second peptide is the SLA epitope, identified via structural modeling. The third peptide is the MMW epitope, also identified through yeast display. The bottom peptide shows the native MART-1 decamer for reference, with the p1 glutamate in parentheses, as DMF5 and most MART-1 TCRs studied recognize both nonameric and decameric MART-1, including p2 anchor-modified variants as studied here.