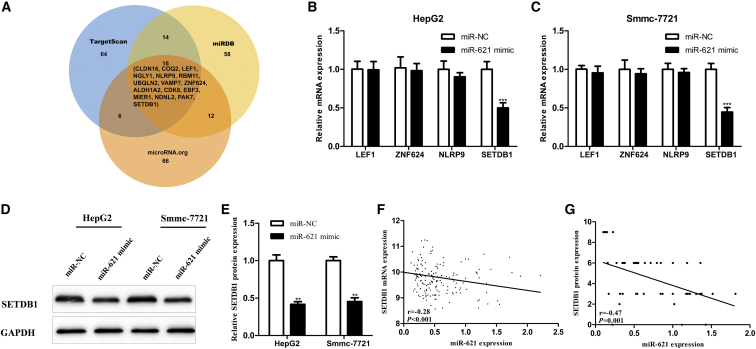
Figure 3.
Screening and Validation of miR-637 Target Genes
(A) TargetScan, miRDB, and microRNA.org were used to predict target genes for miR-621. (B and C) Expressions of LEF1, ZNF624, NLRP9, and SETDB1 mRNA detected by real-time qPCR in HepG2 (B) and Smmc-7721 (C) cells transfected with miR-621 mimic or miR-NC. (D) Western blotting analysis of SETDB1 protein in HepG2 and Smmc-7721 cells transfected with miR-621 mimic or miR-NC. (E) Quantification of SETDB1 protein. (F) The expressions of miR-621 and SETDB1 mRNA in 367 HCC tissues were negatively correlated based on TCGA dataset (r = −0.28, p < 0.001). (G) The expression levels of miR-621 (detected by real-time qPCR) and SETDB1 protein (detected by IHC) in our independent set of 50 HCC tissues were also negatively correlated (r = −0.47, p = 0.001). *p < 0.05, **p < 0.01, ***p < 0.001.