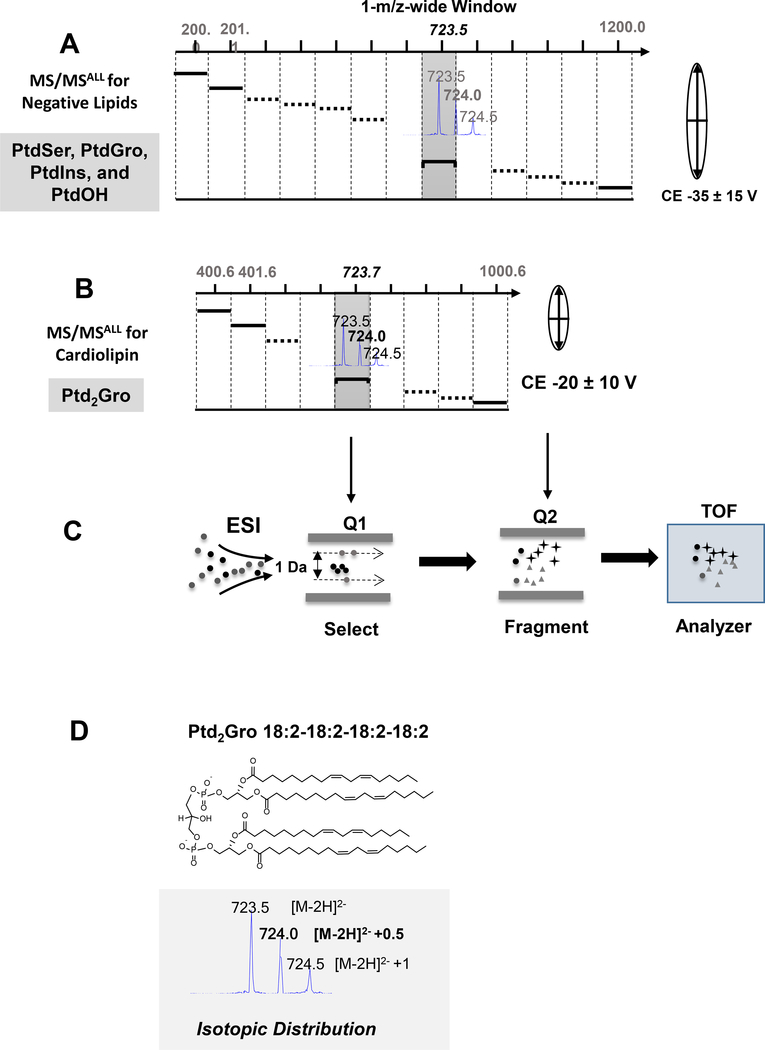
Figure 2. Data independent acquisition MS/MSALL schemes for common negative lipids and cardiolipin.
(A) MS/MSALL for common negative lipids. All precursors are selected in the Q1 at a 1-m/z wide isolation window across 200 to 1200 m/z. Each isolation window is selected to accommodate singly charged negative lipid species with a higher collision energy. (B) MS/MSALL for Ptd2Gro. The Ptd2Gro precursors are selected in Q1 at a 1-m/z wide isolation window across 400 to 1000 m/z with low collision energy and each single adjusted isolated window to fit the doubly charged Ptd2Gro species and isotopic peaks (see inset). (C) ESI-QTOF-MS/MSALL workflow. (D) The molecular structure and isotopic distribution of Ptd2Gro 18:2–18:2–18:2–18:2 under negative ESI conditions. CE, collision energy; ESI, electrospray ionization; PtdOH, phosphatidic acid; PtdGro, phosphatidylglycerol; PtdIns, phosphatidylinositol; PtdSer, phosphatidylserine; Q, quadrupole; TOF, time of flight mass spectrometer.