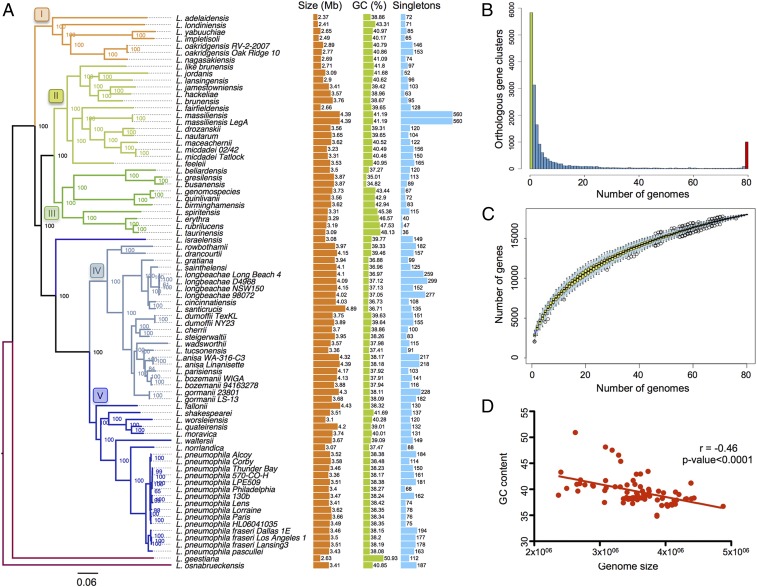
Fig. 1.
The Legionella genomes are diverse in size and gene content. (A) Phylogeny of the genus based on the core genome, genome size, GC content, and number of singletons of each species are depicted. Numbers represent bootstrap values. Branches are colored according to the clade they belong to. Genome size and GC content include plasmids if present in the corresponding species. The number of singletons is based on the results of OrthoMCL (takes into account orthologs and paralogs). Each species has been compared with the others without taking into account strains from the same species to avoid bias due to the number of strains sequenced within a species. (B) Occurrence of genes within the 80 analyzed Legionella genomes. Left end of the x axis (green bar), genes present in a single genome (strain-specific genes; 5,832, ∼32% of the pangenome); right end of the x axis (red bar), genes present in all 80 genomes (core genome; 1,008 genes, ∼6% of the pan-genome). (C) Gene accumulation curve for the total number of proteins of the 80 genomes. (D) Negative correlation between genome size and GC content, indicating high acquisition of foreign genes (Pearson’s correlation coefficient equal to −0.46 with P < 0.0001).