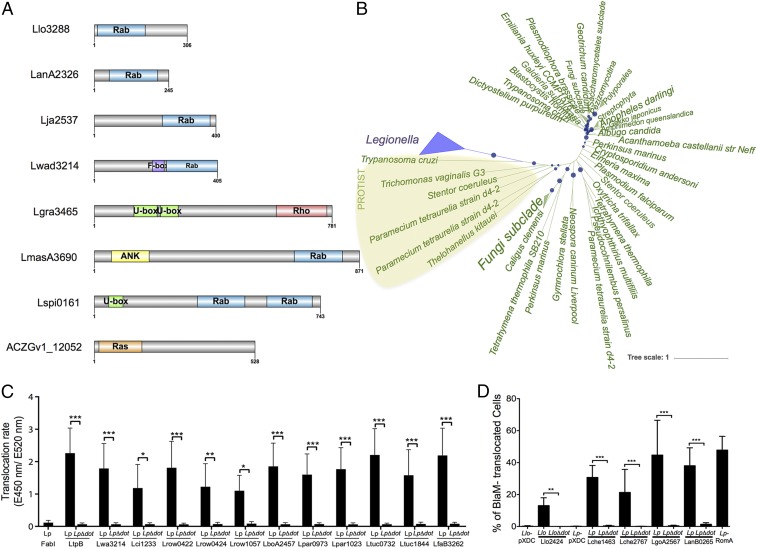
Fig. 3.
Domain organization of small GTPases in Legionella and phylogenetic analyses of the Llo3288 Rab proteins suggest eukaryotic origin. (A) Domain organization of the different small GTPase proteins identified. (B) Unrooted tree of Llo3288 and homologs recruited by blastp constructed using likelihood. Local support values are represented with circles on the corresponding branches, and size of circles is proportional to the values (only local support values of at least 0.7 are shown). (C) Translocation of selected proteins using the beta-lactamase translocation assay and infection of Raw264.7 cells for 1 h with Lp wild type or LpΔdotA expressing BlaM-effector fusions analyzed with a microplate reader. Three independent experiments (n = 9) were done. Statistical significance was determined by two-way ANOVA with multiple comparisons test (*P < 0.05; **P < 0.01; ***P < 0.001). (D) Translocation of selected proteins using the beta-lactamase translocation assay and infection of THP-1 cells at a multiplicity of infection of 50 during 1 h 30 min with Lp and Llo strains before addition of CCF4-AM and analyses by flow cytometry. Histograms show the frequency of BlaM- translocated, blue fluorescence-emitting cells as means ± SD of three independent experiments (n = 12). Statistical significance was determined by Wilcoxon matched pairs test (**P < 0.01; ***P < 0.001). Llo, L. longbeachae wild type; LloΔdot, L. longbeachae ΔdotA; Lp, L. pneumophila wild type; LpΔdot, L. pneumophila ΔdotA.