Significance
Flavin-based electron bifurcation (FBEB) is a long-hidden mode of energetic coupling in which an endergonic electron transfer process is coupled to an exergonic one. The function of the few FBEB complexes described so far is to achieve ferredoxin reduction at the negative redox limit of the biological redox window. Here, a membrane-associated FBEB complex, isolated and characterized from an anaerobic, aromatic compound-degrading bacterium, achieves a redox reaction beyond this limit possibly by two consecutive FBEB events, with reduced ferredoxin serving as donor. The benzene ring-reducing class II benzoyl-CoA reductase has a [Bam(BC)2DEFGHI]2 composition and represents, with 4 W, 2 Se, 6 FAD, and >50 FeS cofactors, one of the most complex electron transfer machineries in nature.
Keywords: metalloenzyme, electron bifurcation, electron transfer, aromatic compound, membrane protein complex
Abstract
Reversible biological electron transfer usually occurs between redox couples at standard redox potentials ranging from +0.8 to −0.5 V. Dearomatizing benzoyl-CoA reductases (BCRs), key enzymes of the globally relevant microbial degradation of aromatic compounds at anoxic sites, catalyze a biological Birch reduction beyond the negative limit of this redox window. The structurally characterized BamBC subunits of class II BCRs accomplish benzene ring reduction at an active-site tungsten cofactor; however, the mechanism and components involved in the energetic coupling of endergonic benzene ring reduction have remained hypothetical. We present a 1-MDa, membrane-associated, Bam[(BC)2DEFGHI]2 complex from the anaerobic bacterium Geobacter metallireducens harboring 4 tungsten, 4 zinc, 2 selenocysteines, 6 FAD, and >50 FeS cofactors. The results suggest that class II BCRs catalyze electron transfer to the aromatic ring, yielding a cyclic 1,5-dienoyl-CoA via two flavin-based electron bifurcation events. This work expands our knowledge of energetic couplings in biology by high-molecular-mass electron bifurcating machineries.
Reversible electron transfer between biological donors/acceptors is generally limited to a redox window between +0.8 and −0.5 V. At the negative limit, reduced ferredoxin (Fd) is the most potent natural electron carrier for biological redox reactions with E′(Fd/Fd–) ≈ −0.5 V, assuming 95% of cellular Fd is in the reduced state. There are only a few biologically relevant redox reactions operating at E°′ values negative enough to reduce Fd: aldehyde:Fd oxidoreductases (E°′ ≈ −580 mV), CO dehydrogenases (E°′ ≈ −520 mV), or 2-oxoacid:Fd oxidoreductases (E°′ ≈ −500 mV). Fd reduction may be achieved with NADH (E°′ = −320 mV) or H2 (E°′ = −414 mV) by membrane-bound NADH:Fd oxidoreductases (Rnf complexes) or by energy-converting hydrogenases (Ech complexes), each driving endergonic Fd reduction by an ion motive force (1–3).
Only 10 y ago, a novel mode of energetic coupling was discovered in which the endergonic reduction of Fd by NADH was coupled to the exergonic reduction of crotonyl-CoA to butyryl-CoA (E°′ = −10 mV) by the same donor, referred to as flavin-based electron bifurcation (FBEB) (4, 5). Meanwhile, a number of FBEB complexes have been described in which the electron bifurcating flavin is first reduced by two electrons from a midpotential donor, followed by two consecutive one-electron transfer steps to a high- and a low-potential acceptor, the latter always representing Fd or flavodoxin (1, 2, 6–11). The biological function of FBEB-driven Fd reduction is either to conserve energy (e.g., via Rnf complexes) or to generate an electron donor for low-potential redox processes (e.g., CO2, H+, or NADP+ reduction).
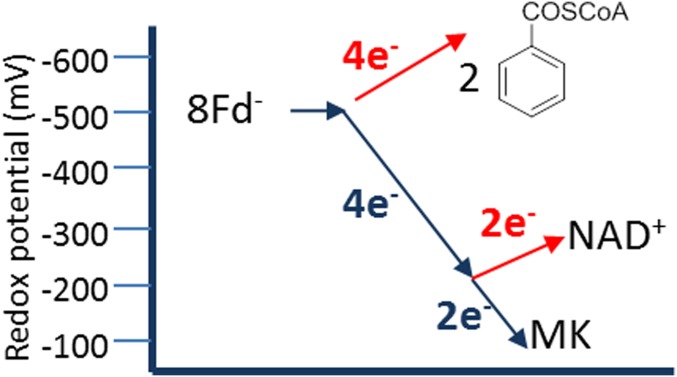
Anaerobic bacteria that use aromatic compounds as growth substrates depend on an enzymatic reaction at a redox potential significantly below that of the Fd/Fd– couple—that is, the reductive dearomatization of the central intermediate benzoyl-CoA to cyclohexa-1,5-diene-1-carboxyl-CoA (1,5-dienoyl-CoA) at E°′ = −622 mV (Fig. 1) (12, 13). These bacteria are important for the global bioremediation of monocyclic aromatic compounds, many of which are harmful to the environment or human health (e.g., benzene, toluene, ethylbenzene, and xylenes). In organic synthesis, aromatic ring reduction to a cyclic diene is accomplished by the Birch reduction, depending on alkali metals dissolved in ammonia as electron donors, on alcohol as a proton donor, and on cryogenic temperatures (14). Considering these conditions, it is remarkable that two totally nonrelated classes of dearomatizing benzoyl-CoA reductases (BCRs) were identified in anaerobic bacteria that reduce their aromatic substrate in a Birch-like manner (12, 15). The soluble class I BCRs contain three [4Fe-4S] cluster and couple benzoyl-CoA reduction by reduced Fd to a stoichiometric ATP hydrolysis (16, 17), whereas the class II BCRs achieve reductive dearomatization at a tungstopterin cofactor in an ATP-independent manner (18, 19). Structural and computational studies with the active-site Bam(BC)2 subunits of class II BCRs from the Fe(III)-respiring deltaproteobacterium Geobacter metallireducens containing the active-site W cofactor and four [4Fe-4S] clusters enabled initial insight into the mechanism of ATP-independent biological Birch reduction (20, 21). However, the components and mechanisms of the underlying energetic coupling have remained hypothetical.
Fig. 1.
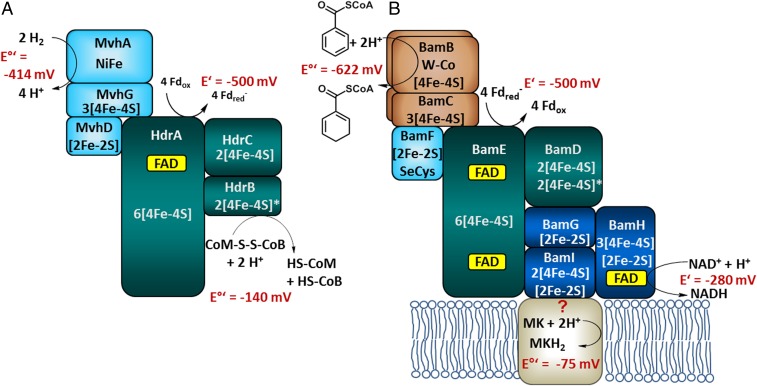
Experimentally verified electron bifurcating MvhAGD–HdrABC complex from methanogenic archaea (A) and the putative membrane-associated class II BCR complex in aromatic compound-degrading bacteria (B). For class II BCRs, it has previously been postulated that endergonic electron transfer from Fd– to benzoyl-CoA is driven by the endergonic reduction of NAD+ by Fd– (18, 22), whereas proteomic analyses (28) and results from this work suggest that a so-far-unknown MK binding component is additionally involved; as a result, the originally proposed electron bifurcation scenario needs to be modified (see Fig. 5). Significant similarities are indicated by identical colors. The BamBC cofactors have been verified experimentally, whereas those of the BamDEFGHI components are based on conserved binding motifs. [4Fe-4S]* indicate noncubane [4Fe-4S] clusters (see also Fig. 3).
The bamBC genes are organized in a cluster together with six further genes (bamDEFGHI), the putative products of which were proposed to be involved in endergonic electron transfer from the likely donor, reduced Fd, to the Bam(BC)2 components (22). The putative gene products show similarities to soluble NADH-binding components of respiratory complex I (BamGHI, related to NuoEFG, Escherichia coli notification) and to components of the electron bifurcating heterodisulfide reductase/hydrogenase complex, HdrABC–MvhAGD, from methanogenic archaea (BamDEF, related to the HdrABC and MvhD) (Fig. 1). The latter couples the endergonic reduction of Fd by H2 to the reduction of the CoM-S-S-CoB heterodisulfide (E°′ = −140 mV) by the same donor via FBEB (23). Meanwhile, a few other enzyme complexes involving HdrA-like components have been shown or proposed to be involved in FBEB processes (24–27). These findings led to the hypothesis that enzymatic dearomatization by class II BCRs is driven by an unprecedented FBEB process in which Fd, in its reduced form, functions as a midpotential donor instead of low-potential acceptor. NAD+ was proposed to serve as second, high-potential acceptor (Fig. 1). With E′ = −500 mV (Fd/Fd–) and E′ = −280 mV (NAD+/NADH), class II BCR reaction was originally hypothesized to proceed as follows (reaction 1) (15, 18):
| [1] |
However, a differential membrane proteome analysis identified components of class II BCR predominantly in the membrane fraction (28), and the reverse methyl viologen oxidation by 1,5-dienoyl-CoA [1,5-dienoyl-CoA oxidoreductase (DCO) activity] was always found (to a significant extent) membrane bound (18), both of which suggest that menaquinone (MK) could serve as an additional/alternative high-potential acceptor (reaction 2 and Fig. 1, with E°′ [MK/MKH2] = −75 mV):
| [2] |
If indeed both NAD+ and MK act as high-potential acceptors, two distinguishable FBEB events would be required to accomplish benzoyl-CoA reduction by Fd–. Here, we developed a strategy for the isolation and characterization of a prototypical 1-MDa, membrane-bound class II BCR complex that drives endergonic benzene ring reduction at the negative biological redox limit by, most likely, two FBEB events.
Results
Homologous Production and Enrichment of Class II BCR Complexes.
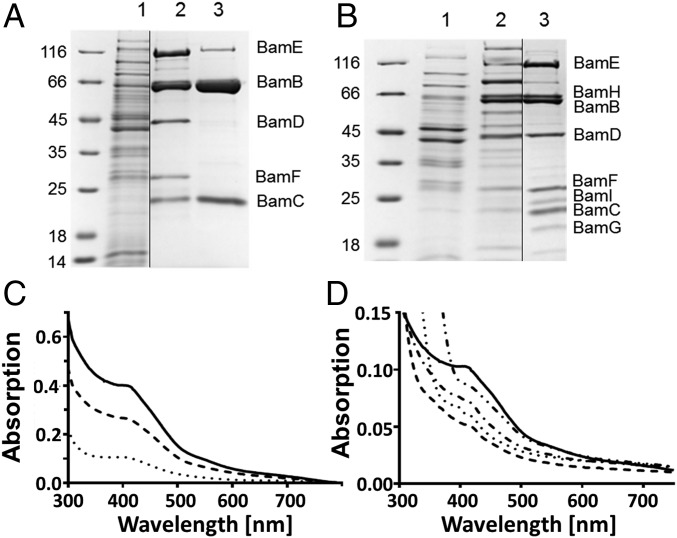
To enrich the hypothesized BamBCDEFGHI complex, we homologously produced BamB with a C-terminal Strep-tag II (BamBStrep) in G. metallireducens pGM2078e1 based on previously established anaerobic expression systems in Geobacter sulfurreducens (29) and G. metallireducens (30). From extracts of this strain, a mixed BamBStrepBC2 complex was enriched after two chromatographic steps (Fig. 2A). Obviously, the plasmid-encoded BamBStrep exchanged with the genomically encoded BamB. The C-terminal tag showed no effect on Bam(BC)2 dimerization and specific DCO activity (Table 1). SDS/PAGE analyses of enriched Twin-Strep-tagged BamB2xStrep revealed the presence of BamB2xStrepBWTC2 and Bam(B2xStrepC)2 complexes in an almost 1:1 ratio (SI Appendix, Fig. S1).
Fig. 2.
Enrichment and UV–visible spectra of different class II BCR (sub)complexes from G. metallireducens. (A) SDS/PAGE analysis of enriched Bam(BStrepC)2 and Bam[(BStrepC)2DEF]2. Lanes: 1, cell extract; 2, fraction eluting at ∼750 kDa; 3, fraction eluting at ∼180 kDa during gel filtration. (B) SDS/PAGE analysis of enriched Bam[(BC)2DEFGHI]2. Lanes: 1, cell extract; 2, after DEAE Sepharose chromatography; 3, after gel filtration. (C) UV–visible spectra normalized to 1 µM BamB concentration of (sub)complexes as isolated: Bam[(BC)2DEFGHI]2 (—), Bam[(BStrepC)2DEF]2 (- - -), and Bam(BStrepC)2 (⋅⋅⋅⋅). (D) UV–visible spectra of 1 µM oxidized Bam[(BC)2DEFGHI]2 (—); reduced with 50 µM NADH (⋅⋅-⋅⋅-); with 50 µM 2-oxoglutarate, 50 μM CoA, 0.2 μmol min−1 KGORTaro, and 0.5 µM Fd (-⋅-⋅); with 50 µM sodium dithionite (⋅⋅⋅⋅); with 50 µM 1,5-dienoyl-CoA (- - -).
Table 1.
Enrichment, catalytic, and molecular properties of different Bam-(sub)complexes of class II BCRs
| Property | Complex | |||
| Subunit architecture | (BC)2* | (BStrepC)2 | [(BStrepC)2DEF]2 | [(BC)2DEFGHI]2 |
| Enrichment of DCO activity, fold | 115 | 74 | 37 | 28 |
| Yield of DCO activity, % | 18 | 25 | 17 | 4.2 |
| Theoretical molecular mass, kDa | 190 | 200 | 758 | 984 |
| Determined molecular mass, kDa | 185 ± 10 | 188 ± 8 | 784 ± 107† | ∼1000† |
| Metal/FAD, mol/mol | ||||
| Fe | 30.4 ± 1.2 | 33.6 | 136 ± 18 | 184 ± 3.8 |
| W | 1.8 | 1.9 | 4.2 ± 0.2 | 4 ± 0.4 |
| Zn | 2.4 | 1.2 | 4.6 ± 0.2 | 5.2 ± 0.4 |
| Se | <0.1 | ND | 1.8 ± 0.4 | 2 ± 0.2 |
| FAD | ND | ND | 4 ± 0.2 | 5.8 ± 0.2 |
| Specific DCO activity, µmol min−1 mg−1 | 68 | 49.9 | 24.6 | 23.9 |
| Km (1,5-dienoyl-CoA)/NADH, µM | 24 ± 4/ND | 19 ± 5/ND | ND/ND | ND/69 ± 12 |
ND, not determined.
Values for the Bam(BC)2 complex were previously determined (19).
Masses >700 kDa are outside the accurate fractionation range of the Superdex 200 gel filtration column used.
The exchange of BamBStrep with BamBWT in Bam(BC)2 complexes motivated us to capture a high-molecular-mass class II BCR complex by omitting a KCl treatment step (19). Using this approach, an ∼750-kDa class II BCR complex was enriched by Strep-Tactin affinity chromatography and gel filtration and was composed of five protein bands migrating at 115, 65, 43, 28, and 21 kDa during SDS/PAGE (Fig. 2A); mass spectrometric analysis identified them as BamBCDEF (Gmet_2083-2087) from G. metallireducens (SI Appendix, Table S1). The DCO activity of Bam(BStrepC)2 was enriched 74-fold, with a yield of 25%, and the one of BamBStrepCDEF was enriched 37-fold, with a yield of 17% (Table 1). The molecular mass and the stoichiometry of subunits on SDS polyacrylamide gels after densitometric analyses clearly pointed to a Bam[(BStrepC)2DEF]2 composition.
The putative BamGHI subunits with high similarities to soluble components of respiratory complex I were missing in the BamBStrep captured complex. Because it is known that the integrity of soluble components of respiratory complex I is optimal at slightly acidic pH (31), a modified strategy was developed to enrich a probable WT BamBCDEFGHI complex from G. metallireducens by adjusting buffers to pH 6. During the enrichment, only fractions that exhibited both DCO and NADH:benzyl viologen oxidoreductase (NOR) activities were considered; the latter activity is characteristic for soluble components of respiratory complex I. Using anion-exchange chromatography and gel filtration, a high-molecular-mass enzyme complex at around 1 MDa was identified in which DCO was 28-fold enriched and NOR activity was 25-fold enriched (Table 1). The apparent low yield of DCO activity (4.2%) can at least partially be assigned to removal of low-molecular BamBC components that were obviously not part of the complex (see below); the low yield of NOR activity (3.8%) is explained by the separation from enzymes exhibiting unspecific NADH oxidizing activities (e.g., soluble components of complex I). SDS/PAGE analysis of the high-molecular-mass enzyme fractions revealed the enrichment of eight protein bands (Fig. 2B). Next to the BamBCDEF components, additional bands migrating around 67, 25, and 19 kDa were identified by mass spectrometry (MS) as the BamGHI components (SI Appendix, Table S1). Densitometric analysis of protein band intensities and the estimated molecular mass are in line with a Bam[(BC)2DEFGHI]2 composition of the complex, with a theoretical molecular mass of around 950 kDa. Taking into account the metal and organic cofactor content, the native molecular mass of the entire complex is estimated to be between 980 and 990 kDa (Table 1).
Metals and Flavin Cofactors.
The previously characterized active-site Bam(BC)2 complex binds one W, one Zn, and four [4Fe-4S] clusters per BamBC unit (19, 21). The BamDEFGHI components identified here are predicted to contain numerous additional redox cofactor domains that are conserved in all class II BCRs. Based on conserved cofactor-binding motifs in the structurally characterized heterodisulfide reductase/hydrogenase HdrABC–MvhADG complex from methanogens (32) and the E. coli NuoEFG components of respiratory complex I (33), the assignment of cofactors to individual class II BCR subunits was carried out.
BamDEF.
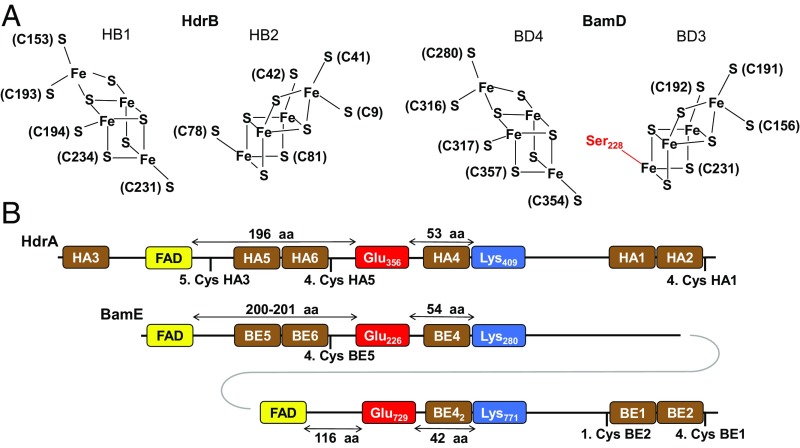
BamD can be regarded as a fusion of the HdrBC components of heterodisulfide reductases from methanogens that harbor two cubane (HC1 and HC2 in HdrC) and two noncubane (HB1 and HB2 in HdrB) [4Fe-4S] clusters (32) (Fig. 1). The latter two represent the active-site cofactors where the CoM-S-S-CoB heterodisulfide is reduced. In BamD, all cysteines of the four clusters are conserved, with the exception of Cys78 coordinating one of the two noncubane clusters in Methanothermococcus thermolithotrophicus; it is replaced by Ser228 in BamD (Fig. 3A and SI Appendix, Table S2). BamE is homologous to HdrA that harbors the electron bifurcation FAD and six [4Fe-4S] clusters (HA1 to HA6) (32). In BamE, HA3 is missing, whereas the domain harboring HA4 and the bifurcating FAD is duplicated (Fig. 3B and SI Appendix, Table S3). The distinguishing sequence signature of the electron bifurcating FAD includes the conserved Glu226/Glu729 and Lys280/Lys771 (notification from BamE) (32). The duplication of the flavin/HA4 binding domain was verified in all 23 examined BamEs from aromatic compound-degrading anaerobes, suggesting that the duplication of the electron bifurcating domain is highly conserved in class II BCRs (SI Appendix, Table S4). BamF, homologous to the hydrogenase MvhD subunit, is predicted to harbor a cysteine-ligated [2Fe-2S] cluster (32). Another conserved cysteine (Cys16 in MvhD) forms a disulfide with one of the cluster-ligating cysteines in the oxidized state; it is replaced by a selenocysteine in all BamFs (SI Appendix, Table S5) and explains the Se dependency during growth with aromatics (22, 34).
Fig. 3.
Conserved cofactor binding modules/motifs in HdrB/BamD and HdrA/BamE. Amino acid (aa) numbering of HdrAB from M. thermolithotrophicus (11) and BamDE from G. metallireducens is shown. (A) Cysteines coordinating noncubane [4Fe-4S] clusters involved in disulfide reduction in HdrB, and the conserved residues found in BamD. (B) Relative arrangement of conserved [4Fe-4S] cluster and FAD-binding motifs in HdrA/BamE; the conserved glutamic acid and lysine residues, typical for electron bifurcating FAD, are shown. HA1 to HA6: [4Fe-4S] clusters in HdrA; BE1 to BE6: [4Fe-4S] clusters in BamE. The FAD-binding domain including BE4 is duplicated in all class II BCRs.
BamGHI.
BamH shows similarities to NuoF from respiratory complex I, with a typical flavin-binding Rossmann fold (V/IxGxGxxGxxxG/A). It is predicted to bind three [4Fe-4S] clusters and a [2Fe-2S] cluster in an additional NuoE-like N-terminal domain (33). BamG, homologous to NuoE, putatively binds another [2Fe-2S] cluster. Finally, the NuoG-like BamI shows motifs for binding two of the four [4Fe-4S] cluster-containing motifs (33) plus an additional [2Fe-2S]-binding thioredoxin-like domain. In summary, the BamDEFGHI components are predicted to harbor 15 [4Fe-4S] and four [2Fe-2S] clusters [one selenocysteine and three flavins (two flavins in BamE and one in BamH)]. Together with the four [4Fe-4S] clusters present in BamBC, this gives a theoretical number of 46 [4Fe-4S] clusters and eight [2Fe-2S] clusters accounting for 200 Fe, two Se, and six flavins per Bam[(BC)2DEFGHI]2 complex.
Inductively coupled plasma MS analysis of Bam[(BC)2DEFGHI]2 identified 184 ± 3.8 Fe per complex, representing more than 92% of the theoretical expected Fe content. In addition, nearly stoichiometric amounts of four W, four Zn (both bound to BamB), and two Se (bound to BamF) per complex were determined, strongly confirming the Bam[(BC)2DEFGHI]2 composition with the theoretically assumed metal/Se content (Table 1). Moreover, HPLC-based flavin cofactor analysis revealed 5.8 ± 0.2 FAD per Bam[(BC)2DEFGHI]2 and 4.0 ± 0.2 mol FAD for Bam[(BStrepC)2DEF]2, which is in full agreement with the predicted binding of two FAD in BamE and one in BamH.
Reduction of Cofactors.
In the as-isolated state, the Bam(BC)2, Bam[(BStrepC)2DEF]2, and Bam[(BC)2DEFGHI]2 complexes exhibited characteristic UV–visible absorbance spectra of fully oxidized FeS proteins, with a shoulder between 400 and 450 nm (Fig. 2C; for extinction coefficients, see SI Appendix, Table S6). In accordance with the estimated number of FeS clusters in the individual complexes, the absorbance of this shoulder was only 67% in Bam[(BC)2DEF]2 and 25% in Bam(BC)2, compared with Bam[(BC)2DEFGHI]2. Using 1,5-dienoyl-CoA as reductant, a spectrum of a virtually completely reduced FeS enzyme was observed, referred to as 100% reduction (Fig. 2D). This finding supports the integrity of the Bam[(BC)2DEFGHI]2 complex that transferred electrons from the 1,5-dienoyl-CoA–reduced active-site W cofactor to the redox centers of the entire complex. With dithionite, reduction was around 83% compared with that of 1,5-dienoyl-CoA (Table S6). To reduce the BCR complex with the proposed natural electron donor, Fd was isolated from G. metallireducens cells grown with benzoate in three chromatographic steps; enriched α-ketoglutarate:Fd oxidoreductase from Thauera aromatica (KGORTaro) served as Fd-reducing enzyme (35). The ready and complete reduction of the two [4Fe-4S] clusters containing Fd to Fd2– by KGORTaro in the presence of excess α-ketoglutarate and CoA was verified by UV–visible spectroscopy (SI Appendix, Fig. S2). Using this system, reduction of class II BCR FeS clusters was observed, albeit only to around 60% compared with that of 1,5-dienoyl-CoA (Table S6). Finally, NADH was tested as a possible BamH binding electron donor. Indeed, bleaching of the spectrum was also observed with 50 µM NADH but only to around 12%, suggesting that only a small fraction of the BamGHI-bound FeS clusters were reduced. The results indicate that next to 1,5-dienoyl-CoA, reduced Fd from G. metallireducens and NADH is a suitable electron donor (or acceptor in the oxidized form) for Bam[(BC)2DEFGHI]2, supporting the hypothesis that it is involved in FBEB-driven benzoyl-CoA reduction.
Electron Transfer Reactions Catalyzed.
The Km value for 1,5-dienoyl-CoA in DCO assays was recently determined (24 µM) (19). The initial rates of benzyl viologen-dependent NADH oxidation studied in the present work (NOR activity) followed Michaelis–Menten kinetics, with a Km of 69 ± 12 µM for NADH; NADPH did not substitute for NADH.
FBEB-driven benzoyl-CoA reduction was tested using Fd (0.5 mM; reduced by KGORTaro, 5 mM 2-oxoglutarate, and 0.5 mM CoA) as donor and using NAD+ as a high-potential acceptor. However, by varying buffers and pH values from 6 to 8, as well as the concentrations of all donors/acceptors, no reduction of benzoyl-CoA to 1,5-dienoyl-CoA was observed. Instead, we identified a 1,5-dienoyl-CoA:Fd oxidoreductase activity (40 nmol min−1 mg−1), whereas NAD+ did not serve as electron acceptor for 1,5-dienoyl-CoA oxidation. In contrast, NAD+ was readily reduced by Fd/KGORTaro (440 nmol min−1 mg−1). These results suggest that electron transfer was possible between reduced Fd and the assumed electron output modules BamB and BamH, but not directly between the output modules, which is in line with the general mechanism of FBEB.
A large number of donor/acceptor combinations were tested using Bam[(BC)2DEFGHI]2 and Bam[(BC)2DEF]2 complexes comprising reduced Fd, Ti(III)-citrate, dithionite, reduced methyl viologen, or NAD(P)H as donors and using NAD(P)+, ferrocenium, dichlorophenolindophenol, ferricyanide, disulfides (cystine, glutathione disulfide), or 1,5-dienoyl-CoA (potentially reduced to 1-monoenoyl-CoA by BamB) as acceptors. Attempts were also carried out with crude extracts from G. metallireducens, including the membrane protein fraction using crotonyl-CoA, NO3−, fumarate, or menadione as electron acceptors. Finally, combinations of two electron acceptors were tested with Bam[(BC)2DEFGHI]2 in the presence of crude extracts (for an overview of combinations tested, see SI Appendix, Table S7). Under no conditions were even traces (≥0.1 µM) of 1,5-dienoyl-CoA observed by ultra-performance LC–electrospray ionization–quadrupole TOF–MS analyses.
Subcellular Localization.
The inability to observe benzoyl-CoA reduction with numerous batches of soluble class II BCR complexes suggests that essential components of the electron bifurcation machinery are missing. The observation that DCO activity was always found up to 40 to 50% in the membrane fractions under mild crude extract preparation (SI Appendix, Table S8) suggested that class II BCRs may interact with so-far-unknown membrane components.
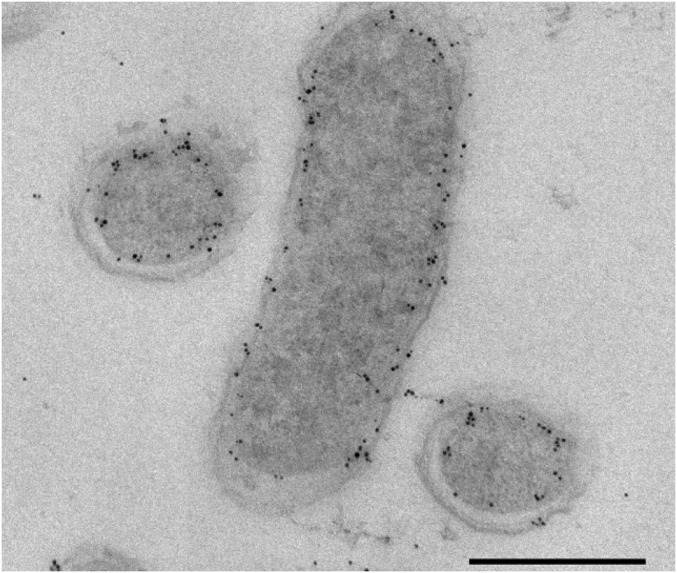
To study the subcellular localization of class II BCRs, polyclonal antibodies raised against BamF (a central component of the Bam[(BC)2DEFGHI]2 complex) were produced. Western blot analyses revealed that BamF is more abundant in the membrane fraction (SI Appendix, Fig. S3), which is in full agreement with a previous membrane proteome analysis of G. metallireducens in which BamF was predominantly found in the membrane fraction (28). To visualize the cellular localization of BamF, immunogold detection of specific anti-BamF antibodies on ultrathin sections was carried out by transmission electron microscopy. Analysis of benzoate-grown G. metallireducens cells showed that the gold particles bound to the anti-BamF antibody were found exclusively localized at the cytoplasmic membrane, leaving little doubt that the Bam[(BC)2DEFGHI]2 complex is membrane bound in vivo (Fig. 4).
Fig. 4.
Immunogold labeling of G. metallireducens cells grown on benzoate with anti-BamF antibodies. The signal of the gold particles, seen as black dots, is localized near the cytoplasmic membrane. (Scale bar, 500 nm.)
Abundance of HdrA-Like Electron Bifurcation Modules.
HdrA from methanogenic archaea can be considered the prototype of a phylogenetically distinct class of electron bifurcating modules. A phylogenetic analysis showed that HdrA-like components are also widely abundant in nonmethanogenic Archaea, Proteobacteria, Firmicutes, and some other prokaryotes (SI Appendix, Fig. S4), indicating that they play an important role in many, so-far-unknown FBEB processes. A phylogenetic subcluster of HdrA-like modules comprises BamE components from obligately anaerobic bacteria that are known to degrade aromatic compounds. Notably, all BamE-like components of this subcluster show the duplication of the FAD-binding domain, suggesting that two electron bifurcation events are essential for class II BCR catalysis.
Discussion
Almost 10 y after the initial isolation of the Bam(BC)2 subcomplex (19) the experimental evidence for the hypothesized high-molecular-mass Bam[(BC)2DEFGHI]2 class II BCR was provided in this work. The exceptional complexity of this class II BCR is reflected by the unprecedented, most probably double FBEB-driven redox reaction catalyzed, in which Fd in its reduced form serves as donor and not as ultimate acceptor, as described for all currently known FBEB processes (1, 2, 7–10). The ready reduction of redox cofactors by reduced Fd from G. metallireducens, NADH, and 1,5-dienoyl-CoA, together with the coenrichment of DCO and NOR activity, initially pointed to an FBEB process, with reduced Fd serving as midpotential donor and with NAD+ serving as a high-potential acceptor and benzoyl-CoA as a low-potential acceptor (Fig. 1B). However, a number of findings of this work now argue for the involvement of additional membranous components: (i) benzoyl-CoA reduction coupled to NAD+ reduction was never observed, even despite the almost complete redox cofactor occupation; (ii) immunogold-labeling studies evidenced that BamF, adapting the active-site Bam(BC)2 to the core bifurcation module, is almost exclusively found at the cytoplasmic membrane; and (iii) in all class II BCRs, the FBEB domain of the BamE subunit is duplicated. All these findings point to a double FBEB-driven benzoyl-CoA reduction involving four electron input/output modules, with MK serving as the most likely membranous high-potential electron acceptor, next to NAD+. It is evident that a double FBEB-driven process could neither be monitored with the soluble Bam[(BC)2DEFGHI]2 complex nor with crude extracts, because in the latter exergonic shortcut electron transfer reactions are barely avoidable. Thus, the identification reconstitution of membranous MK binding components in artificial liposomes together with the Bam[(BC)2DEFGHI]2 components will be the challenge of future work. The results obtained at this stage are in agreement with a double FBEB scenario discussed in the following, where a Fd–-like compound serves as donor for both endergonic benzoyl-CoA reduction and endergonic NAD+/MK reduction (reaction 3 and Fig. 5).
| [3] |
G. metallireducens contains genes encoding KGOR, producing six Fd– per benzoyl-CoA oxidized in the TCA cycle (one benzoyl-CoA yields three acetyl-CoA, SI Appendix, Fig. S5). Considering that this organism also uses Fd– for acetyl-CoA assimilation via pyruvate synthase, KGOR alone is likely to be insufficient to maintain high Fd–/Fd ratios. But its genome contains benzoate-induced genes putatively encoding an electron transferring flavoprotein (ETF) with a [4Fe-4S] cluster binding Fd domain adjacent to a gene for a putative membrane-bound, MK dependent oxidoreductase (Gmet_2065-2067) (28). Similar to the FixABCX complex from Azotobacter vinelandii (36) or Rhodopseudomonas palustris (37), these genes are likely to encode a membrane-bound FBEB machinery in which the endergonic electron transfer from NADH to the Fd domain in ETF is coupled to the exergonic reduction of MK. Thus, the combined action of KGOR and ETF/MK reductase guarantees a high Fd–/Fd ratio.
Fig. 5.
Schematic scenario for double FBEB processes driving benzoyl-CoA reduction by class II BCRs. Red and dark blue arrows indicate endergonic and exergonic electron transfer reactions, respectively.
The second FBEB process is considered to be involved in electron transfer from the second electron bifurcating FAD to NAD+ and MK. In this context, it is noteworthy that the special sequence signature of the two noncubane [4Fe-4S] clusters in HdrB involved in CoM-S-S-CoB reduction at E°′ > −200 mV (38) are conserved in BamD (Fig. 3). Notably, in HdrB, such clusters are reduced by single electrons to achieve a formal two-electron reduction of the heterodisulfide (32). In nonmethanogens lacking the CoM-S-S-CoB, a proteinogenic Cys-S-S-Cys may serve as intermediary electron acceptor, which, after reduction, would be reoxidized by the second FAD (E°′ ≈ −200 mV), finally bifurcating electrons to reduce NAD+ (endergonic branch) and MK (exergonic branch). In such a scenario, energy would be conserved compared with a system that would use only MK as a high-potential acceptor.
The identification of HdrA as a key component of class II BCRs demonstrates the globally important role of this electron bifurcation module in nature in addition to its originally described function in methanogenesis. Its occurrence in nonmethanogenic and nonaromatic compound-degrading prokaryotes indicates that there are a great number of HdrA-like, FBEB-dependent redox machineries that await discovery (SI Appendix, Fig. S4). Among HdrA-containing FBEB, the 1-MDa class II BCR represents an unprecedented metalloenzyme redox machinery—probably one of the most complex ones in nature.
Materials and Methods
Experimental procedures for cultivation of cells, protein expression and enrichment, activity assays, structural and functional analysis of proteins, and immunogold labeling are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Jana Seifert for conducting mass spectrometric analysis of protein samples. This work was funded by the German Research Foundation Grants BO 1565/14-1 and HU 703/2.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819636116/-/DCSupplemental.
References
- 1.Buckel W, Thauer RK. Flavin-based electron bifurcation, a new mechanism of biological energy coupling. Chem Rev. 2018;118:3862–3886. doi: 10.1021/acs.chemrev.7b00707. [DOI] [PubMed] [Google Scholar]
- 2.Buckel W, Thauer RK. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na(+) translocating ferredoxin oxidation. Biochim Biophys Acta. 2013;1827:94–113. doi: 10.1016/j.bbabio.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Fuchs G. Alternative pathways of carbon dioxide fixation: Insights into the early evolution of life? Annu Rev Microbiol. 2011;65:631–658. doi: 10.1146/annurev-micro-090110-102801. [DOI] [PubMed] [Google Scholar]
- 4.Li F, et al. Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J Bacteriol. 2008;190:843–850. doi: 10.1128/JB.01417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herrmann G, Jayamani E, Mai G, Buckel W. Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J Bacteriol. 2008;190:784–791. doi: 10.1128/JB.01422-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan Z, Ferry JG. Electron bifurcation and confurcation in methanogenesis and reverse methanogenesis. Front Microbiol. 2018;9:1322. doi: 10.3389/fmicb.2018.01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters JW, et al. A new era for electron bifurcation. Curr Opin Chem Biol. 2018;47:32–38. doi: 10.1016/j.cbpa.2018.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Müller V, Chowdhury NP, Basen M. Electron bifurcation: A long-hidden energy-coupling mechanism. Annu Rev Microbiol. 2018;72:331–353. doi: 10.1146/annurev-micro-090816-093440. [DOI] [PubMed] [Google Scholar]
- 9.Buckel W, Thauer RK. Flavin-based electron bifurcation, ferredoxin, flavodoxin, and anaerobic respiration with protons (Ech) or NAD+ (Rnf) as electron acceptors: A historical review. Front Microbiol. 2018;9:401. doi: 10.3389/fmicb.2018.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baymann F, et al. On the natural history of flavin-based electron bifurcation. Front Microbiol. 2018;9:1357. doi: 10.3389/fmicb.2018.01357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan Z, Wang M, Ferry JG. A ferredoxin- and F420H2-dependent, electron-bifurcating, heterodisulfide reductase with homologs in the domains bacteria and archaea. MBio. 2017;8:e02285-16. doi: 10.1128/mBio.02285-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs G, Boll M, Heider J. Microbial degradation of aromatic compounds - from one strategy to four. Nat Rev Microbiol. 2011;9:803–816. doi: 10.1038/nrmicro2652. [DOI] [PubMed] [Google Scholar]
- 13.Kung JW, et al. Reversible biological Birch reduction at an extremely low redox potential. J Am Chem Soc. 2010;132:9850–9856. doi: 10.1021/ja103448u. [DOI] [PubMed] [Google Scholar]
- 14.Birch AJ. The birch reduction in organic synthesis. Pure Appl Chem. 1996;68:553–556. [Google Scholar]
- 15.Boll M, Löffler C, Morris BE, Kung JW. Anaerobic degradation of homocyclic aromatic compounds via arylcarboxyl-coenzyme a esters: Organisms, strategies and key enzymes. Environ Microbiol. 2014;16:612–627. doi: 10.1111/1462-2920.12328. [DOI] [PubMed] [Google Scholar]
- 16.Unciuleac M, Boll M. Mechanism of ATP-driven electron transfer catalyzed by the benzene ring-reducing enzyme benzoyl-CoA reductase. Proc Natl Acad Sci USA. 2001;98:13619–13624. doi: 10.1073/pnas.241375598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boll M, Fuchs G. Benzoyl-coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. ATP dependence of the reaction, purification and some properties of the enzyme from Thauera aromatica strain K172. Eur J Biochem. 1995;234:921–933. doi: 10.1111/j.1432-1033.1995.921_a.x. [DOI] [PubMed] [Google Scholar]
- 18.Löffler C, et al. Occurrence, genes and expression of the W/Se-containing class II benzoyl-coenzyme A reductases in anaerobic bacteria. Environ Microbiol. 2011;13:696–709. doi: 10.1111/j.1462-2920.2010.02374.x. [DOI] [PubMed] [Google Scholar]
- 19.Kung JW, et al. Identification and characterization of the tungsten-containing class of benzoyl-coenzyme A reductases. Proc Natl Acad Sci USA. 2009;106:17687–17692. doi: 10.1073/pnas.0905073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Culka M, Huwiler SG, Boll M, Ullmann GM. Breaking benzene aromaticity-computational insights into the mechanism of the tungsten-containing benzoyl-CoA reductase. J Am Chem Soc. 2017;139:14488–14500. doi: 10.1021/jacs.7b07012. [DOI] [PubMed] [Google Scholar]
- 21.Weinert T, et al. Structural basis of enzymatic benzene ring reduction. Nat Chem Biol. 2015;11:586–591. doi: 10.1038/nchembio.1849. [DOI] [PubMed] [Google Scholar]
- 22.Wischgoll S, et al. Gene clusters involved in anaerobic benzoate degradation of Geobacter metallireducens. Mol Microbiol. 2005;58:1238–1252. doi: 10.1111/j.1365-2958.2005.04909.x. [DOI] [PubMed] [Google Scholar]
- 23.Kaster AK, Moll J, Parey K, Thauer RK. Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc Natl Acad Sci USA. 2011;108:2981–2986. doi: 10.1073/pnas.1016761108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mock J, Wang S, Huang H, Kahnt J, Thauer RK. Evidence for a hexaheteromeric methylenetetrahydrofolate reductase in Moorella thermoacetica. J Bacteriol. 2014;196:3303–3314. doi: 10.1128/JB.01839-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramos AR, et al. The FlxABCD-HdrABC proteins correspond to a novel NADH dehydrogenase/heterodisulfide reductase widespread in anaerobic bacteria and involved in ethanol metabolism in Desulfovibrio vulgaris Hildenborough. Environ Microbiol. 2015;17:2288–2305. doi: 10.1111/1462-2920.12689. [DOI] [PubMed] [Google Scholar]
- 26.Cao X, et al. Lipoate-binding proteins and specific lipoate-protein ligases in microbial sulfur oxidation reveal an atpyical role for an old cofactor. eLife. 2018;7:e37439. doi: 10.7554/eLife.37439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milton RD, Ruth JC, Deutzmann JS, Spormann AM. Methanococcus maripaludis employs three functional heterodisulfide reductase complexes for flavin-based electron bifurcation using hydrogen and formate. Biochemistry. 2018;57:4848–4857. doi: 10.1021/acs.biochem.8b00662. [DOI] [PubMed] [Google Scholar]
- 28.Heintz D, et al. Differential membrane proteome analysis reveals novel proteins involved in the degradation of aromatic compounds in Geobacter metallireducens. Mol Cell Proteomics. 2009;8:2159–2169. doi: 10.1074/mcp.M900061-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coppi MV, Leang C, Sandler SJ, Lovley DR. Development of a genetic system for Geobacter sulfurreducens. Appl Environ Microbiol. 2001;67:3180–3187. doi: 10.1128/AEM.67.7.3180-3187.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oberender J, Kung JW, Seifert J, von Bergen M, Boll M. Identification and characterization of a succinyl-coenzyme A (CoA):benzoate CoA transferase in Geobacter metallireducens. J Bacteriol. 2012;194:2501–2508. doi: 10.1128/JB.00306-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leif H, Sled VD, Ohnishi T, Weiss H, Friedrich T. Isolation and characterization of the proton-translocating NADH: Ubiquinone oxidoreductase from Escherichia coli. Eur J Biochem. 1995;230:538–548. doi: 10.1111/j.1432-1033.1995.tb20594.x. [DOI] [PubMed] [Google Scholar]
- 32.Wagner T, Koch J, Ermler U, Shima S. Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction. Science. 2017;357:699–703. doi: 10.1126/science.aan0425. [DOI] [PubMed] [Google Scholar]
- 33.Gnandt E, Dörner K, Strampraad MFJ, de Vries S, Friedrich T. The multitude of iron-sulfur clusters in respiratory complex I. Biochim Biophys Acta. 2016;1857:1068–1072. doi: 10.1016/j.bbabio.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 34.Peters F, Rother M, Boll M. Selenocysteine-containing proteins in anaerobic benzoate metabolism of Desulfococcus multivorans. J Bacteriol. 2004;186:2156–2163. doi: 10.1128/JB.186.7.2156-2163.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dörner E, Boll M. Properties of 2-oxoglutarate:ferredoxin oxidoreductase from Thauera aromatica and its role in enzymatic reduction of the aromatic ring. J Bacteriol. 2002;184:3975–3983. doi: 10.1128/JB.184.14.3975-3983.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ledbetter RN, et al. The electron bifurcating FixABCX protein complex from Azotobacter vinelandii: Generation of low-potential reducing equivalents for nitrogenase catalysis. Biochemistry. 2017;56:4177–4190. doi: 10.1021/acs.biochem.7b00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duan HD, et al. Distinct properties underlie flavin-based electron bifurcation in a novel electron transfer flavoprotein FixAB from Rhodopseudomonas palustris. J Biol Chem. 2018;293:4688–4701. doi: 10.1074/jbc.RA117.000707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madadi-Kahkesh S, et al. A paramagnetic species with unique EPR characteristics in the active site of heterodisulfide reductase from methanogenic archaea. Eur J Biochem. 2001;268:2566–2577. doi: 10.1046/j.1432-1327.2001.02141.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.