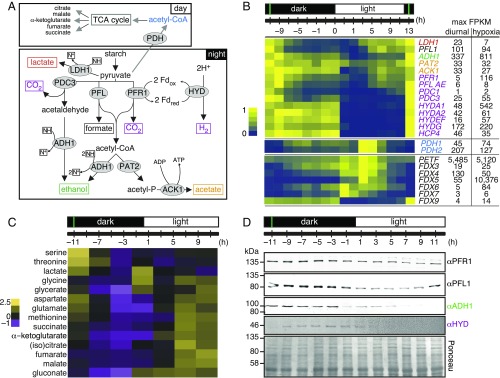
Fig. 6.
Chlamydomonas cells use anaerobic routes for handling pyruvate. (A) Key pyruvate metabolism pathways according to refs. 36 and 46. Final fermentation products are shown in boxes and enzymes in gray ellipses. Abbreviations: ACK1, acetate kinase 1; ADH1, acetaldehyde/alcohol dehydrogenase; HYDA, [Fe–Fe]-hydrogenase; LDH1, d-lactate dehydrogenase; PAT2, phosphate acetyltransferase 2; PFL1, pyruvate formate lyase; PFR1, pyruvate ferredoxin oxidoreductase. N+, NAD+; NH, NADH. (B) Normalized expression of fermentation genes listed, shown as a heatmap. Numbers on the right side indicate maximum FPKM values in our samples (diurnal) and in cells grown under dark hypoxia for 6 h (34). (C) Changes in water-soluble metabolites over the diurnal cycle, analyzed by GC-MS. Results are shown as a heatmap of z-score normalized abundance for metabolites that changed significantly over the diurnal cycle. (D) Fermentation enzymes are more abundant in the dark. Total protein samples were separated by denaturing SDS/PAGE, followed by immune-detection with antibodies raised against PFR1, PFL, ADH, and HYDA1+2. Equal protein amounts were loaded and confirmed with Ponceau S stain. All data are shown as average ± SD (n = 3). The immune-detection was performed at least twice on independent samples. The green vertical lines indicate the timing of cell division.