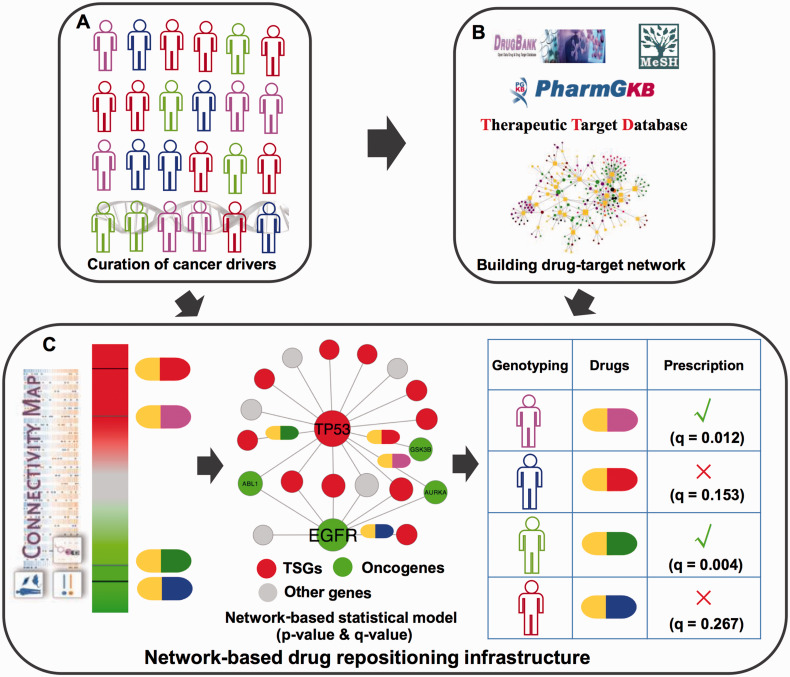
Figure 1:
Diagram of network-based infrastructure for prioritizing potential druggable targets and searching for the existing drugs targeting significantly mutated genes (SMGs) in cancer genomes. This infrastructure is designed for better development of newly targeted cancer therapies. (A) Manual curation of SMGs discovered in large-scale cancer genome data generated from The Cancer Genome Atlas and public domains (see the Materials and Methods section). (B) Searching for new potential druggable anticancer targets via mapping the curated SMGs in A into drug-target interaction network collected from public databases: DrugBank, PharmGKB, and Therapeutic Target Database. (C) Development of a network-based drug repositioning approach to prioritize new potential anticancer indications for existing drugs. It incorporates drug-gene signatures from the Connectivity Map, the manually curated SMGs, and the protein-protein interaction network using a statistical approach (see the Materials and Methods section). TSGs: tumor suppressor genes.