Abstract
Quantification of co-migrating paraproteins in the beta-region presents an ongoing challenge for laboratories performing serum protein electrophoresis. The between-laboratory variation may impact patient care if the patient uses different pathology services during plasma cell dyscrasia monitoring. To identify the practical difficulties and determine the extent of agreement in the reporting of beta-migrating paraproteins in Australia and New Zealand (NZ), sample exchanges were conducted in five Australian states and in NZ in early 2018. This study has highlighted the variation in quantification and reporting of beta-migrating paraproteins which could potentially affect patient monitoring and management.
Introduction
Quantification of paraprotein migrating in the beta-region presents an ongoing challenge for laboratories performing serum protein electrophoresis. At the Australasian Association of Clinical Biochemists (AACB) and Royal College of Pathologists of Australasia Quality Assurance Program (RCPAQAP) Proteins Workshop held in Melbourne in September 2017 participants discussed ways to best quantify and report beta-migrating paraproteins that would result in greater consistency of results between laboratories. Currently, there is no accurate method of quantifying beta-migrating paraproteins either by serum protein electrophoresis (SPEP), by total immunoglobulin (Ig) assays or using heavy/light chain assays. Paraprotein concentrations may include polyclonal Ig(s) or other normal co-migrating proteins such as transferrin and C3 complement, resulting in their overestimation by densitometry or immunometric methods. The between-laboratory variation in quantification and reporting of beta-migrating paraproteins may impact patient care if the patient uses different pathology services with different laboratory SPEP methods during disease response monitoring.1
The 2012 recommendations for standardised quantification and reporting of paraproteins are due for revision; in particular, the quantification and reporting of beta-migrating paraproteins.2 Information regarding the between-laboratory variation of paraprotein values by SPEP and Ig assays and current laboratory electrophoresis practices are required before the recommendations can be updated. The ultimate aim is to better harmonise the quantification and reporting of paraproteins by Australian and NZ laboratories when monitoring disease response.3
To identify the practical problems and level of agreement in the reporting of beta-migrating paraproteins in Australia and NZ, sample exchanges were conducted in five Australian states and in NZ in early 2018. The aim of the sample exchange was to assess variation in practice for the quantification and reporting of beta-migrating paraproteins and also assess possibilities for improved harmonisation; for example, using the serum total Ig concentration (e.g. IgG, IgA or IgM) or the total beta-region plus paraprotein as the paraprotein measurand for the monitoring of response.
Materials and Methods
Laboratories in five Australian states and NZ were invited to participate in the sample exchange project in February 2018. States in Australia and NZ had local coordinators who prepared samples. Sufficient volumes of serum containing mainly beta-migrating paraproteins (the Queensland sample exchange contained one sample with a gamma-migrating paraprotein) of IgA isotype but also IgG and IgM types were sourced from left-over routine patient samples by the coordinators. Samples were de-identified prior to dispatch in aliquots to other local or NZ laboratories on ice or dry-ice. The samples were not spiked or pooled from multiple sera. A minimum of four samples with varying concentrations were distributed within five Australian states and NZ. On receipt of samples, laboratories were requested to store them at −20 °C or −80 °C until analysis.
The isotype of the paraprotein was provided by the coordinator. The laboratories were invited to quantify the paraproteins and report paraprotein concentration using their routine practice and also measure the involved Ig using immunonephelometric assay (INA) or immunoturbidimetric assay (ITA). The participating laboratories from Victoria were also requested to measure ‘total beta + paraprotein’ by densitometry on SPEP for each sample.
A spreadsheet for the collection of results was distributed to each group of participants on which the serum total protein and albumin concentrations were provided using the coordinating laboratory’s methods. In addition to entering the paraprotein concentration and total Ig, participants were asked to state their SPEP method and the platform used to quantify immunoglobulins in their laboratory.
Data Analysis
The results were compared between laboratories in five Australian states and in NZ using the mean concentration of the paraprotein or total involved Ig, calculated for each group of local Australian laboratories (numbers varied from 2 to 8) and NZ laboratories (N=10). In general, paraprotein concentrations were reported in whole numbers whereas total Ig concentrations were reported to one decimal place. The coefficient of variation (CV) was calculated and compared for each sample.
The paraprotein concentration displayed in the figures and tables reflect the various ways that laboratories quantify and report paraproteins using different SPEP methods. Paraprotein concentrations were determined by: ‘perpendicular drop’ (PD); ‘tangent skimming’ (TS); ‘total beta + paraprotein’; ‘total beta-1 or beta-2 + paraprotein’; ‘corrected perpendicular drop’ (cPD) where the quantified area is sometimes narrowed in an attempt to compensate for the included normal proteins, possibly guided by immunosubtraction; or ‘total beta minus a pre-determined concentration of normal beta globulins’ (Figures 1 and 2). The advantages and disadvantages of different gating methods have been described by Keren and Schroeder.4
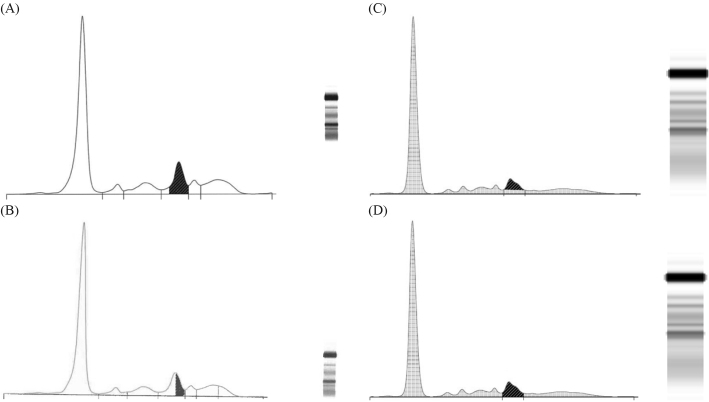
Figure 1.
(A) Densitometric scan of sample 4 analysed by New Zealand (NZ) laboratory 4 using perpendicular drop gating and Capillarys® capillary zone electrophoresis (CZE) methods. IgA lambda was reported as 8 g/L (hatched area). Note the paraprotein is in the beta-1 zone. (B) Densitometric scan of sample 4 analysed by NZ laboratory 3 using corrected perpendicular drop gating and Capillarys® CZE methods. IgA lambda was reported as 4 g/L (hatched area). Note the paraprotein is in the beta-1 zone. (C) Densitometric scan of sample 4 analysed using tangent skimming gating and high-resolution agarose gel electrophoresis (HR AGE) methods. IgA lambda would be reported as 4 g/L (hatched area). Note the paraprotein is in the beta-2 zone. (D) Densitometric scan of sample 4 analysed by NZ laboratory 9 using perpendicular drop gating and HR AGE methods. IgA Lambda was reported as 6 g/L (hatched area). Note the paraprotein is in the beta-2 zone.
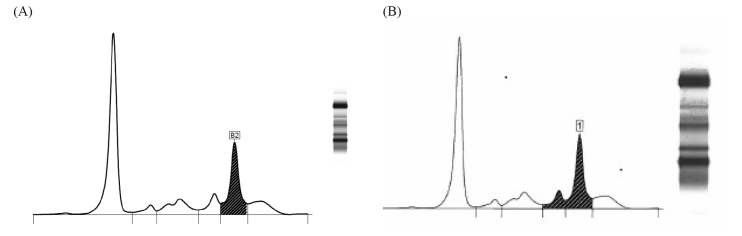
Figure 2.
(A) Densitometric scan of sample 2 analysed by Queensland laboratory 3 using Capillarys® capillary zone electrophoresis (CZE) method. IgA kappa paraprotein concentration was reported as the ‘total beta-2 + paraprotein concentration’ = 19 g/L (hatched area). (B) Densitometric scan of sample 2 analysed by Queensland laboratory 1 using Capillarys® CZE method. IgA kappa paraprotein concentration was reported as the ‘total beta + paraprotein concentration’ = 26 g/L (hatched area).
Results
Thirty-one laboratories from both public and private pathology services returned results. The respondent details are shown in Table 1.
Table 1.
Sample exchange participants.
| State/Country | No. of laboratories invited | No. of participants | No. of samples in exchange |
|---|---|---|---|
| New South Wales | 10 | 3 | 4 |
| New Zealand | 10 | 10 | 4 |
| Queensland | 4 | 4 | 10 |
| South Australia | 2 | 2 | 8 |
| Victoria | 8 | 8 | 4 |
| Western Australia | 4 | 4 | 5 |
Information relating to the SPEP method and Ig quantification method in each of the sample exchange groups is listed in Table 2. Of the 31 participating laboratories, 58% used capillary zone electrophoresis (CZE) and 42% agarose gel electrophoresis (AGE). The Ig quantification method varied between participating laboratories.
Table 2.
Summary of methods used in sample exchange (expressed as number of laboratories in each state or country).
| NSW | NZ | Qld | SA | Vic | WA | |
|---|---|---|---|---|---|---|
| Serum protein electrophoresis method | ||||||
| Sebia Capillarys® | 2 | 7 | 3 | 4 | 1 | |
| Sebia agarose gel system | 1 | 2 | 1 | 2 | 4 | |
| Helena capillary zone electrophoresis | 1 | |||||
| Helena agarose gel system | 2 | |||||
| Lab-based agarose gel system | 1 | |||||
| Ig quantification method/platform | ||||||
| Abbott Architect c/ci/i series | 1 | 1 | 3 | 2 | ||
| Beckman Coulter AU5800 | 1 | |||||
| Beckman Coulter IMMAGE/IMMAGE 800 | 1 | 1 | 2 | |||
| Binding Site SPA PLUS | 1 | 1 | ||||
| Binding Site OPTILITE | 1 | |||||
| Roche Diagnostics Cobas c501/c502 | 3 | 1 | ||||
| Roche Diagnostics Cobas c701/c702 | 1 | 1 | 1 | |||
| Siemens ADVIA 2400 | 1 | 1 | 1 | 1 | ||
| Siemens Diagnostics (Dade Behring) BN II | 1 | 1 | 2 | |||
| Siemens Diagnostics (Dade Behring) ProSpec | 1 | |||||
We compared the reported paraprotein concentrations and total Ig concentrations between laboratories for five Australian states and NZ. Results are shown in Tables 3–8. The comparison of paraprotein and total Ig concentrations between NZ and Victorian laboratories are shown in Figures 3 and 4. The gating methods used to quantify the paraprotein in NZ and Victorian laboratories are included in Tables 4 and 8. In addition, the laboratories in Victoria measured and provided ‘total beta + paraprotein’ concentration on all samples for this study (Table 9 and Figure 4).
Table 3.
Comparison of reported paraprotein and total Ig concentrations (g/L) between New South Wales laboratories.
| Sample 1 | Sample 2 | Sample 3 | Sample 4 | |||||
|---|---|---|---|---|---|---|---|---|
| Paraprotein | Total IgM | Paraprotein | Total IgG | Paraprotein | Total IgA | Paraprotein | Total IgA | |
| NSW Lab 1 | 25 | 34.2 | 1 | 6.4 | * | 5.9 | 11 | 14.2 |
| NSW Lab 2 | 28 | 33.5 | 3 | 6.6 | 6 | 6.5 | 16 | 13.9 |
| NSW Lab 3 | 24 | 36.1 | 5 | - | 5 | 7.7 | 12 | 14.3 |
| Mean (g/L) | 26 | 35 | 3 | 6.5 | 6 | 6.7 | 13 | 14.1 |
| SD (g/L) | 2.1 | 1.3 | 2.0 | 0.1 | 0.7 | 0.9 | 2.8 | 0.2 |
| CV% | 8.2 | 3.9 | 66.7 | 2.2 | 12.9 | 13.7 | 21.8 | 1.5 |
two co-migrating IgA bands not individually quantifiable
Table 4.
Comparison of paraprotein and total Ig concentrations (g/L) between New Zealand laboratories.
| Assay | Gating | Sample 1 | Sample 2 | Sample 3 | Sample 4 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Paraprotein * | Total IgA | Paraprotein | Total IgA | Paraprotein | Total IgA | Paraprotein | Total IgA | |||
| NZ Lab 1 | IH AGE | PD | 32 (47) | 43.9 | 9 | 10.2 | 28 | 26.2 | 10 | 6.4 |
| NZ Lab 2 | C CZE | PD | 33 (46) | 39.8 | 10 | 9.9 | 25 | 23.9 | 9 | 5.9 |
| NZ Lab 3 | C CZE | PD | 46 | 38.5 | 9 | 9.7 | 25 | 25.2 | 4 | 6.3 |
| NZ Lab 4 | C CZE | PD | 47 | 41.2 | 9 | 10.5 | 25 | 25.3 | 8 | 6.3 |
| NZ Lab 5 | C CZE | PD | 46 | 37.3 | 9 | 9.7 | 25 | 24.2 | 7 | 6.0 |
| NZ Lab 6 | C CZE | TS | 37 | 34.5 | 4 | 10.3 | 16 | 26.0 | 4 | 5.8 |
| NZ Lab 7 | C CZE | PD | 46 | 36.2 | 10 | 9.6 | 26 | 23.1 | 8 | 5.7 |
| NZ Lab 8 | AGE | PD | 44 | 36.7 | 8 | 8.7 | 25 | 22.8 | 9 | 5.8 |
| NZ Lab 9 | HR AGE | PD | 41 | 45.7 | 6 | 9.2 | 23 | 29.1 | 6 | 5.9 |
| NZ Lab 10 | C CZE | PD | 47 | 41.0 | 8 | 9.5 | 25 | 25.7 | 9 | 6.1 |
| Mean (g/L) | 42 | 39.5 | 8 | 9.7 | 24 | 25.2 | 7 | 6.0 | ||
| SD (g/L) | 5.9 | 3.5 | 1.9 | 0.5 | 3.2 | 1.8 | 2.1 | 0.2 | ||
| CV% | 14.0 (7.3) | 9.0 | 22.9 | 5.5 | 13.0 | 7.3 | 28.6 | 4.1 | ||
| CV% (C CZE All) | 13.0 (7.9) | 24.6 | 14.6 | 30.9 | ||||||
| CV% (C CZE PD only) | 12.4 (1.1) | 8.2 | 1.6 | 24.9 | ||||||
Sample 1 has two peaks.
The value for the first peak only was reported by Labs 1 & 2. Results in brackets reflect the combined concentration of two peaks. AGE, agarose gel electrophoresis; C, Capillarys®; CZE, capillary zone electrophoresis; HR, high resolution; PD; perpendicular drop; IH; in-house
Table 5.
Comparison of reported paraprotein and total Ig concentrations (g/L) between Queensland laboratories.
| Sample 1 | Sample 2 | Sample 3 | Sample 4 | Sample 5 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Paraprotein* | Total IgA | Paraprotein | Total IgA | Paraprotein | Total IgA | Paraprotein | Total IgA | Paraprotein | Total IgA | |
| Qld Lab 1 | 79 | 60.0 | 26 | 19.0 | 30 | 30.0 | 15 | 11.0 | 14 | 18.0 |
| Qld Lab 2 | 74 | 59.0 | 34 | 15.0 | 22 | 31.0 | 7 | 6.0 | 10 | 15.0 |
| Qld Lab 3 | 76 | 53.0 | 19 | 12.5 | 28 | 24.0 | 8 | 6.7 | 12 | 13.0 |
| Qld Lab 4 | 76 | ND | 21 | 14.0 | 30 | ND | 14 | 7.0 | 16 | ND |
| Mean (g/L) | 76 | 57.3 | 25 | 15.1 | 28 | 28.3 | 11 | 7.7 | 13 | 15.3 |
| SD (g/L) | 2.1 | 3.8 | 6.7 | 2.8 | 3.8 | 3.8 | 4.1 | 2.3 | 2.6 | 2.5 |
| CV% | 2.7 | 6.6 | 26.7 | 18.4 | 13.8 | 13.4 | 37.1 | 29.4 | 19.9 | 16.4 |
|
| ||||||||||
| Sample 6 | Sample 7 | Sample 8 | Sample 9 | Sample 10 | ||||||
| Paraprotein | Total IgG | Paraprotein | Total IgM | Paraprotein | Total IgM | Paraprotein | Total IgG | Paraprotein | Total IgG | |
|
| ||||||||||
| Qld Lab 1 | 11 | 10.0 | 12 | 4.0 | 30 | 36.0 | 48 | 35.0 | 11 | 6.0 |
| Qld Lab 2 | - | 12.0 | 3 | 5.0 | 28 | 26.0 | 40 | 29.0 | 2 | 7.0 |
| Qld Lab 3 | 9 | 11.0 | 7 | 5.0 | 29 | 36.0 | 46 | 40.0 | 4 | 6.0 |
| Qld Lab 4 | 12 | 11.0 | 12 | ND | 32 | ND | 49 | 43.0 | 11 | ND |
| Mean (g/L) | 11 | 11.0 | 9 | 4.7 | 30 | 32.7 | 46 | 36.8 | 7 | 6.3 |
| SD (g/L) | 1.5 | 0.8 | 4.4 | 0.6 | 1.7 | 5.8 | 4.0 | 6.1 | 4.7 | 0.6 |
| CV% | 14.3 | 7.4 | 51.3 | 12.4 | 5.7 | 17.7 | 8.8 | 16.7 | 67.0 | 9.1 |
IgA paraprotein migrated in the gamma region and was used as a gating control;
ND, not done
Table 6.
Comparison of reported paraprotein and total Ig concentrations (g/L) between Western Australian laboratories.
| Sample 1 | Sample 2 | Sample 3 | Sample 4 | Sample 5 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Paraprotein | Total IgM | Paraprotein | Total IgA | Paraprotein | Total IgG | Paraprotein | Total IgA | Paraprotein | Total IgG | |
| WA Lab 1 | 2 | 5.4 | 7 | 3.7 | 4 | 5.0 | 9 | 5.8 | 2 | 8.4 |
| WA Lab 2 | 4 | 4.9 | 4 | 4.1 | 3 | 5.3 | 10 | 6.3 | 1 | 8.4 |
| WA Lab 3 | ND | ND | ND | ND | ND | ND | 7 | ND | ND | ND |
| WA Lab 4 | 3 | 5.0 | * | * | * | * | 7 | 6.1 | 1 | 8.5 |
| Mean (g/L) | 3 | 5.1 | 6 | 3.9 | 4 | 5.2 | 8 | 6.1 | 1 | 8.4 |
| SD (g/L) | 1.0 | 0.3 | 2.1 | 0.3 | 0.7 | 0.2 | 1.5 | 0.3 | 0.6 | 0.1 |
| CV% | 33.3 | 5.2 | 38.6 | 7.3 | 20.2 | 4.1 | 18.2 | 4.1 | 43.3 | 0.7 |
Insufficient sample;
ND, not done
Table 7.
Comparison of reported paraprotein and total Ig concentrations (g/L) between South Australian laboratories.
| Sample 1 | Sample 2 | Sample 3 | Sample 4 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Paraprotein | Total IgA | Paraprotein | Total IgA | Paraprotein | Total IgG | Total IgM | Paraprotein | Total IgA | |
| SA Lab 1 | 8 | 7.9 | 35 | 46.8 | 10 | 14.8 | 9.3 | 5 | 6.5 |
| SA Lab 2 | 8 | 7.6 | 36 | 46.7 | 11 | 16.3 | 9.4 | 7 | 7.2 |
| Mean (g/L) | 8 | 7.8 | 36 | 46.8 | 11 | 15.6 | 9.4 | 6 | 6.9 |
| SD (g/L) | 0.0 | 0.2 | 0.7 | 0.1 | 0.7 | 1.1 | 0.1 | 1.4 | 0.5 |
| CV% | 0.0 | 2.7 | 2.0 | 0.2 | 6.7 | 6.8 | 0.8 | 23.6 | 7.2 |
|
| |||||||||
| Sample 5 | Sample 6 | Sample 7 | Sample 8 | ||||||
| Paraprotein | Total IgA | Paraprotein | Total IgA | Paraprotein | Total IgA | Paraprotein | Total IgA | ||
|
| |||||||||
| SA Lab 1 | 22 | 24.8 | 9 | 6.3 | 9 | 11.1 | 8 | 5.5 | |
| SA Lab 2 | 30 | 26.7 | 9 | 6.8 | 10 | 10.8 | 7 | 5.6 | |
| Mean (g/L) | 26 | 25.8 | 9 | 6.55 | 10 | 11.0 | 8 | 5.6 | |
| SD (g/L) | 5.7 | 1.3 | 0.0 | 0.4 | 0.7 | 0.2 | 0.7 | 0.1 | |
| CV% | 21.8 | 5.2 | 0.0 | 5.4 | 7.4 | 1.9 | 9.4 | 1.3 | |
Sample 3 – two co-migrating bands of IgG and IgM
Table 8.
Comparison of reported paraprotein and total Ig concentrations (g/L) between Victorian laboratories.
| Assay | Gating system for beta-migrating PP | Sample 1 | Sample 2 | Sample 3 | Sample 4 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Paraprotein | Total IgA | Paraprotein | Total IgA | Paraprotein | Total IgA | Paraprotein | Total IgA | |||
| Vic Lab 1 | C CZE | Total β + PP | 9 | 3.5 | 11 | 5.3 | 17 | 9.1 | 54 | 43.4 |
| Vic Lab 2 | HR AGE | NA* | Not detected | - | 4 | 5.2 | 9* | 8.8 | 41* | 40.6 |
| Vic Lab 3 | C CZE | Total β1 or β2 + PP | <1 | 3.5 | 4 | 5.3 | 6 | 10.2 | 51 | 41.5 |
| Vic Lab 4 | HR AGE | Total β1 or β2 + PP | 3 | 3.7 | 2 | 5.6 | 6 | 7.8 | 41 | 46.4 |
| Vic Lab 5 | AGE | Total β + PP - std β value |
2 | 3.4 | 4 | 5.0 | 11 | 9.0 | 59 | 40.3 |
| Vic Lab 6 | AGE | Total β1 or β2 + PP | 6 | 3.5 | 3 | 5.1 | 6 | 8.5 | 51 | 40.8 |
| Vic Lab 7 | C CZE | Total β + PP | 11 | 3.7 | 13 | 6.1 | 18 | 8.3 | 59 | 44.8 |
| Vic Lab 8 | C CZE | Total β1 or β2 + PP or total β + PP | 6 | 3.5 | 5 | 5.3 | 17 | 10.3 | 52 | 42.3 |
| Mean (g/L) | 6 | 3.5 | 6 | 5.4 | 11 | 9.0 | 51 | 42.5 | ||
| SD (g/L) | 3.4 | 0.1 | 4.0 | 0.3 | 5.4 | 0.9 | 7.0 | 2.2 | ||
| CV% | 55.2 | 3.2 | 70.5 | 6.5 | 47.9 | 9.7 | 13.8 | 5.2 | ||
Total Ig by turbidimetry reported;
AGE, agarose gel electrophoresis; C, Capillarys®; CZE, capillary zone electrophoresis; HR, high resolution; PP, paraprotein; NA, not available
Figure 3.
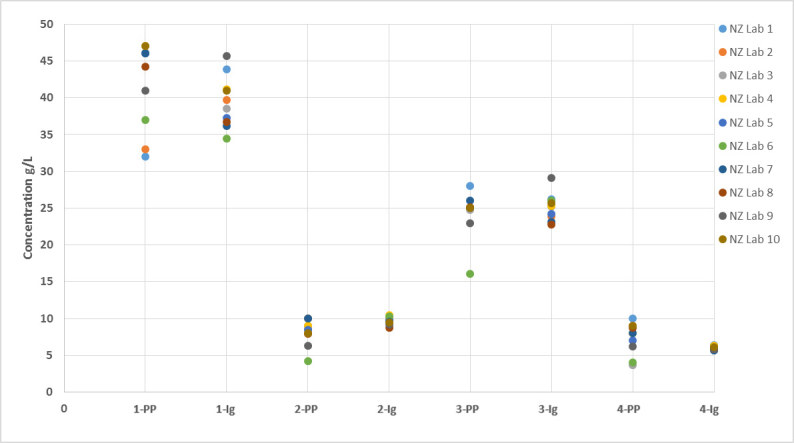
Comparison of paraprotein and total Ig concentrations between NZ laboratories. (The number denotes the sample number; PP – paraprotein concentration; Ig – total Ig concentration.)
Figure 4.
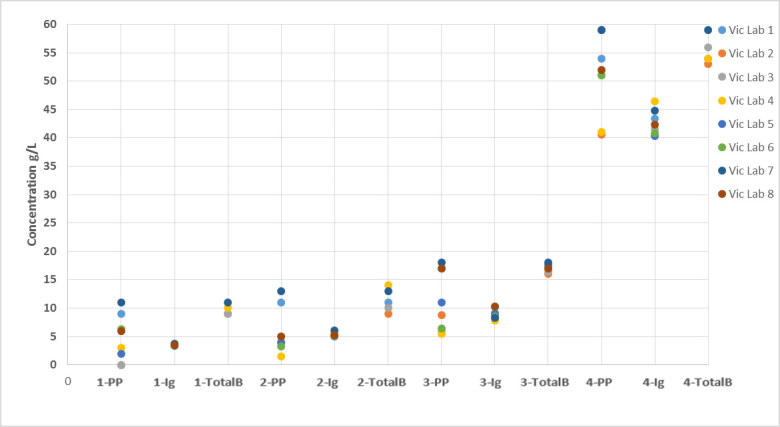
Comparison of paraprotein, total Ig and ‘total beta + paraprotein’ concentrations between Victorian laboratories. (The number denotes the sample number; PP – paraprotein concentration; Ig – total Ig concentration; TotalB – ‘total beta + paraprotein’ concentration.)
Table 9.
Comparison of reported ‘total beta + paraprotein’ concentrations (g/L) between Victorian laboratories.
| Sample 1 | Sample 2 | Sample 3 | Sample 4 | |
|---|---|---|---|---|
| Vic Lab 1 | 9 | 11 | 17 | 54 |
| Vic Lab 2 | Not detected | 9 | 16 | 53 |
| Vic Lab 3 | 9 | 10 | 16.4 | 56 |
| Vic Lab 4 | 10 | 14 | 17.2 | 54 |
| Vic Lab 5 | ND | ND | 17.6 | ND |
| Vic Lab 6 | ND | ND | ND | ND |
| Vic Lab 7 | 11 | 13 | 18 | 59 |
| Vic Lab 8 | ND | ND | 17 | ND |
| Mean (g/L) | 10 | 11 | 17 | 55 |
| SD (g/L) | 1.0 | 2.1 | 0.7 | 2.4 |
| CV% | 9.8 | 18.2 | 4.0 | 4.3 |
ND, not done
Overall, for all participants, quantification of the involved Ig by immunochemical methods performed better than what is reported in the paraprotein field (Tables 3–8). CVs for total Ig vs paraprotein were: 1.5–13.7% vs 8.2–66.7% for NSW, 4.1–9.0% vs 13–28.6% for NZ, 7.4–29.4% vs 5.7–67.0% for Qld, 0.7–7.3% vs 18.2–43.3% for WA, 0.2–7.2% vs 0–23.6% for SA and 3.2–9.7% vs 13.8–70.5% for Vic.
Table 9 shows Victorian laboratories that also reported ‘total beta + paraprotein’. CVs for this quantification method are improved over what is reported in the paraprotein field (4.0–18.2 % vs 13.8–70.5%.
Examples of how the paraprotein gating method influences the reported paraprotein concentration are shown in Figures 1 and 2. Figure 1 shows selected examples from laboratories in NZ participating in the sample exchange. Laboratory 4 separated sample 4 by Capillarys® CZE and quantified the paraprotein by PD. The hatched area represents the IgA lambda paraprotein in the beta-1 zone and includes some underlying normal beta-1 proteins. The result was reported as ‘IgA lambda = 8 g/L’ (Figure 1A). Laboratory 3 also separated the sample by Capillarys® CZE but, using cPD (guided by immunosubtraction) gating method, obtained a paraprotein concentration of 4 g/L (Figure 1B). Using HR AGE, the same paraprotein was located in the beta-2 zone rather than in the beta-1 zone by Capillarys® CZE. Using TS as the gating method, the paraprotein concentration was estimated as 4 g/L (Figure 1C) compared with 6 g/L using PD (Figure 1D) by laboratory 9.
Figure 2 shows automatic gating of serum proteins separated by Capillarys® CZE with an IgA kappa paraprotein located in the beta-2 region from the Queensland sample exchange. In Figure 2A the hatched area represents the IgA kappa paraprotein and other beta-migrating normal proteins. There is no operator manipulation of the scan. The result was reported as ‘total beta-2 + IgA kappa’ = 19 g/L by Queensland laboratory 3 whereas it was reported as ‘total beta + IgA kappa’ = 26 g/L by laboratory 1 which also used Capillarys® CZE (Figure 2B).
Discussion
This sample exchange project aimed to investigate the variation in quantification of paraproteins in the beta-region that co-migrated with normal proteins on SPEP. It also aimed to compare the total Ig measurements of the involved or monoclonal Ig using various INA/ITA methods.
The sample exchange confirmed that the concentration reported in the paraprotein field on a laboratory report has a significant between-laboratory variation, similar to the findings observed in the RCPAQAP Paraprotein Program. The CVs varied, up to 71%, generally depending on the paraprotein concentration, with lower paraprotein concentrations having highest CVs. The extent of between-laboratory variation is different amongst Australian states and NZ. This may reflect differences in the systems used (CZE or AGE), or the variation in the gating methods.
Analysis of the NZ data in particular shows that some of the between-laboratory variation can be accounted for by method-specific migration and resolution differences (e.g. CZE vs AGE), but the impact was small compared to gating methodology (Table 4). These results suggest that the largest contributors to variation in paraprotein quantification between laboratories are the gating method selected (Figure 1) and operator variability when applying the gating method. A study performed by the Mayo Clinic involving 16 laboratories has found a lack of accuracy with both PD and TS (Jillian R. Tate, personal communication) and in the absence of a reference method for paraprotein quantification, it is not possible to assess whether gating using either TS or PD is more accurate. The amount of normal co-migratory beta-region proteins included with the paraprotein quantification will vary depending on which gating methodology is used; therefore gating methodology should ideally be specified with paraprotein quantification. Adopting PD gating across laboratories would be a pragmatic approach to reduce the variation due to gating methodology and is recommended by the Intergroupe Francophone du Myélome (IFM). 5
Reducing between-operator variability when applying the gating method may be more difficult to address. However, using a standard approach across the total beta-region or individual beta-1 or beta-2 regions, including the paraprotein, may improve precision. When a standardised quantification procedure (‘total beta + paraprotein’) was used by the Victorian laboratories, CVs reduced from 14–71% to 4–18% (Tables 8 and 9), suggesting that using the same reporting procedure improves between-laboratory agreement in paraprotein quantification. We did not assess the between-laboratory variation using ‘total beta-1 or total beta-2 + paraprotein’ method during this study. This may also provide for the possibility of improved harmonisation but may also be hampered by the differences in migration and resolution of different SPEP methods and systems.
In general, measurement of the total involved Ig by immunometric methods as a surrogate measure of the monoclonal Ig concentration gave the lowest between-laboratory variation with CVs varying between 0.2 and 29.4%, indicating that different INA/ITA platforms give comparable values for most paraproteins. However, the current reporting practices for quantitative Igs are not harmonised between laboratories as indicated by the RCPAQAP survey which demonstrated that the upper limit of reporting by laboratories is highly variable (range from ~2 g/L to any value).3 More work is required to compare quantitative electrophoresis and immunometric measurements for monoclonal IgG, IgA and IgM to determine harmonised upper limits of reporting for use by all laboratories. As demonstrated by this study, the limiting factor is not the platform but is more likely to be the nature of the paraprotein molecule with IgM paraproteins ‘notorious’ for their overestimation by INA. This does not mean that high values should not be reported but rather that there should be appropriate commenting indicating the measurement uncertainty.2
Overall, opportunities for improved harmonisation of the reported beta-region paraprotein concentrations include the reporting of total involved Ig and/or the reporting of ‘total beta + paraprotein’ using standardised gating as shown by the improved between-laboratory CVs exhibited by these methods. This supports suggestions made at the 2017 Harmonisation Workshop to use total Ig to monitor patient response. For IgA paraproteins, this is also in agreement with the 2016 International Myeloma Working Group consensus criteria for response and minimal residual disease in multiple myeloma which states quantitative Igs are preferred for disease assessments for IgA myelomas.6 However, SPEP is still required for monitoring these patients for response classification e.g. complete response/very good partial response as indicated in clinical guidelines.6–8
Conclusions
This study has highlighted the between-laboratory variation in measurement of beta-migrating paraproteins within Australia and NZ. This variation potentially affects patient care when a patient is being monitored for treatment response. A patient may have their blood collected (and subsequently analysed) at multiple pathology services for convenience, being unaware that there are differences in laboratory reporting of monoclonal proteins measured by SPEP. Similarly, treating clinicians may not be aware of the method differences and this may lead to changes in treatment decisions, misclassification of response and difficulty in accessing clinical trials or expensive anti-myeloma therapies.
The paraprotein sample exchange has shown that the between-laboratory variation is more likely to be related to how laboratories quantify and report paraproteins. Dejoie et al. note in their IFM recommendations for uniform interpretation of serum and urine protein electrophoresis in multiple myeloma diagnosis and follow-up that “the value of the peak concentration does not have a prognostic value itself, but is fundamental for response-to-treatment evaluation based on the % of variation of the peak. The initial concentration on diagnosis is the reference value for response to treatment assessment.”5 Opportunities for improved harmonisation of the reported beta-region paraprotein concentrations include the reporting of total involved Ig and/or the reporting of ‘total beta + paraprotein’. Further studies would be required to assess the suitability of other quantification methods including ‘total beta-1 or total beta-2 + paraprotein’. Standardising the gating methodology to PD where applied, as recommended by IFM, would also be expected to reduce variability in paraprotein quantification.
Acknowledgements
Thanks go to all participating laboratories which included:
NSW – Immunology Laboratory, Royal Prince Alfred Hospital; Immunology Laboratory, John Hunter Hospital; Immunology Laboratory, St Vincent’s Hospital, Sydney.
New Zealand – Canterbury Health Laboratories, Christchurch; Canterbury Southern Community Laboratories, Christchurch; LabPLUS, Auckland; Labtests, Auckland; Medlab Central, Palmerston North; Middlemore Hospital Laboratory, Auckland; Pathlab Waikato, Hamilton; Southern Community Laboratories, Dunedin; Taranaki Medlab, New Plymouth; Waikato Hospital Laboratory, Hamilton.
Queensland – Mater Pathology, Mater Hospital, South Brisbane; Biochemistry Dept, Queensland Medical Laboratories, Murarrie; Chemical Pathology, Pathology Queensland, Royal Brisbane and Women’s Hospital, Herston; Immunology Dept, Sullivan Nicolaides Pathology, Bowen Hills.
South Australia – SA Pathology - Frome Rd, Adelaide; Australian Clinical Labs, Wayville.
Victoria – Alfred Pathology Service, Alfred Health, Melbourne; Austin Pathology, Austin Health, Heidelberg; Dorevitch Pathology, Heidelberg; Melbourne Pathology, Collingwood; Monash Pathology, Monash Health, Clayton; Melbourne Health Pathology, Royal Melbourne Hospital, Parkville; Pathology Dept, Peter MacCallum Cancer Centre, Parkville; St Vincent’s Pathology, St. Vincent’s Hospital, Fitzroy.
Western Australia – PathWest, QEII Medical Centre, Nedlands; PathWest Fiona Stanley Hospital, Murdoch; Western Diagnostic Pathology, Myaree; Clinipath, Osborne Park.
Footnotes
Competing Interests: None declared.
References
- 1.Tate JR, Keren DF, Mollee P. A global call to arms for clinical laboratories - Harmonised quantification and reporting of monoclonal proteins. Clin Biochem. 2018;51:4–9. doi: 10.1016/j.clinbiochem.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Tate J, Caldwell G, Daly J, Gillis D, Jenkins M, Jovanovich S, et al. Recommendations for standardized reporting of protein electrophoresis in Australia and New Zealand. Ann Clin Biochem. 2012;49:242–56. doi: 10.1258/acb.2011.011158. [DOI] [PubMed] [Google Scholar]
- 3.Wijeratne N, Tate JR, Wienholt L, Mollee P. Report of the survey conducted by RCPAQAP on current practice for paraprotein and serum free light chain measurement and reporting: a need for harmonisation. Clin Biochem Rev. 2019;40:31–42. [PMC free article] [PubMed] [Google Scholar]
- 4.Keren DF, Schroeder L. Challenges of measuring monoclonal proteins in serum. Clin Chem Lab Med. 2016;54:947–61. doi: 10.1515/cclm-2015-0862. [DOI] [PubMed] [Google Scholar]
- 5.Dejoie T, Lakomy D, Caillon H, Pegourié B, Decaux O. IFM (Intergroupe francophone du myélome) recommendations for uniform interpretation of serum and urine protein electrophoresis in multiple myeloma diagnosis and follow-up. Ann Biol Clin (Paris) 2016;74:429–41. doi: 10.1684/abc.2016.1166. [DOI] [PubMed] [Google Scholar]
- 6.Kumar S, Paiva B, Anderson KC, Durie B, Landgren O, Moreau P, et al. International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 2016;17:e328–46. doi: 10.1016/S1470-2045(16)30206-6. [DOI] [PubMed] [Google Scholar]
- 7.Comenzo RL, Reece D, Palladini G, Seldin D, Sanchorawala V, Landau H, et al. Consensus guidelines for the conduct and reporting of clinical trials in systemic light-chain amyloidosis. Leukemia. 2012;26:2317–25. doi: 10.1038/leu.2012.100. [DOI] [PubMed] [Google Scholar]
- 8.Owen RG, Kyle RA, Stone MJ, Rawstron AC, Leblond V, Merlini G, et al. Response assessment in Waldenström macroglobulinaemia: update from the VIth International Workshop. Br J Haematol. 2013;160:171–6. doi: 10.1111/bjh.12102. [DOI] [PubMed] [Google Scholar]