Fig 1.
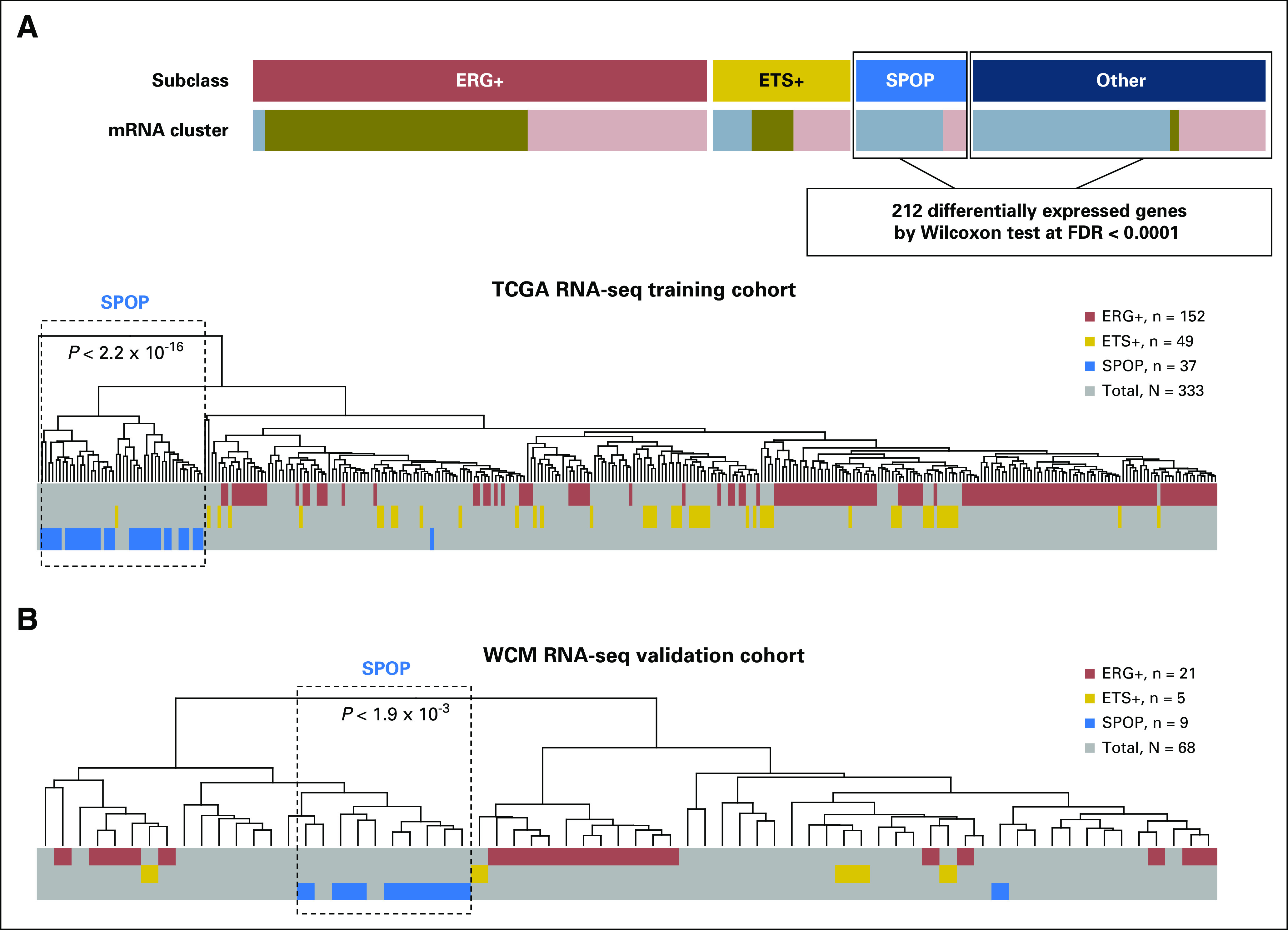
SPOP mutant transcriptional signature. (A) SPOP mutant transcriptional signature that included 212 differentially expressed genes between SPOP mutant and wild-type samples from The Cancer Genome Atlas (TCGA) non-ETS fusion RNA sequencing (RNA-seq) data. The signature was generated from the TCGA study and tested back in the TCGA training cohort. Significant enrichment of SPOP mutant cases was based on hierarchical clustering of 333 TCGA prostate cancer samples. Different colors represent molecular subclasses from genomic and transcriptomic annotations. (B) Significant enrichment of SPOP mutant case from 68 Weill Cornell Medicine (WCM) prostate cancer samples with SPOP mutant transcriptional signature on the basis of hierarchical clustering. ERG, ERG-fusion position; ETS: other ETS fusion positive; FDR, false-discovery rate.
