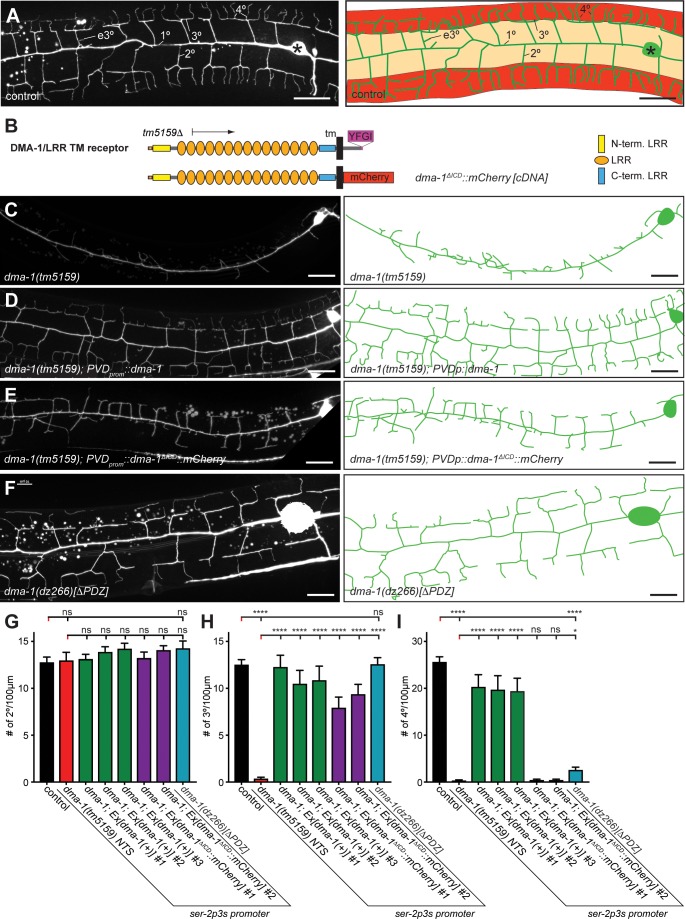
Figure 1. The intracellular domain of DMA-1/LRR-TM is required for higher order branching of PVD somatosensory dendrites.
(A) Fluorescent images of PVD (left panels) and schematics (right panels) of wild-type control animals. PVD is visualized by the wdIs52 [F49H12.4p::GFP] transgene in all panels. 1°, 2°, 3°, 4°, and ectopic 3° (e3°) dendrites are indicated. Anterior is to the left and dorsal is up in all panels, scale bars indicate 20 µm. (B) Schematics of the DMA-1/LRR-TM protein and a variant used in transgenic rescue experiments (dma-1(ΔICD)). A PDZ-binding site (YFGI) at the extreme C-terminus of DMA-1/LRR-TM is indicated in lilac. The predicted deletion from the tm5159 deletion allele is shown. (C-F) Fluorescent images of PVD (left panels) and schematics (right panels) of the genotypes indicated. Scale bar indicates 20 µm. (G) Quantification of 2°, 3°, and 4° branch numbers per 100 µm anterior to the PVD cell body. Data for three and two independent transgenic lines for the dma-1 wild type cDNA or the dma-1(ΔICD), respectively, are shown next to the data for the dma-1(dz266[∆PDZ]) allele. The data for dma-1(tm5159) NTS are nontransgenic siblings of a representative transgenic line. For raw data see Figure 1—source data 1. Data are represented as mean ± SEM. Statistical comparisons were performed using one-sided ANOVA with Sidak’s correction. Statistical significance is indicated (ns, not significant; ****, p < 0.0001). n = 20 animals per genotype.