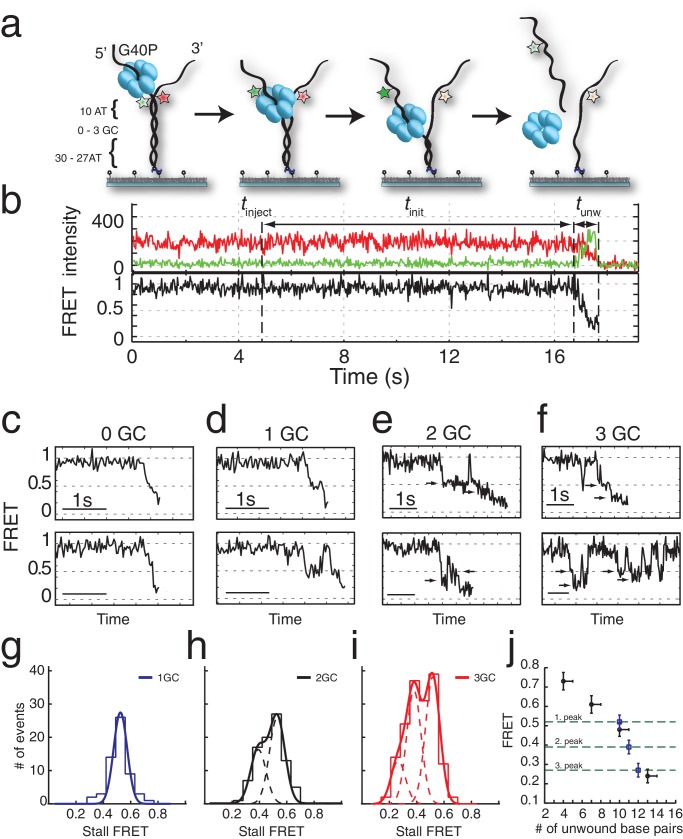
Figure 1. G40P unwinds DNA in one base pair steps und slips backwards.
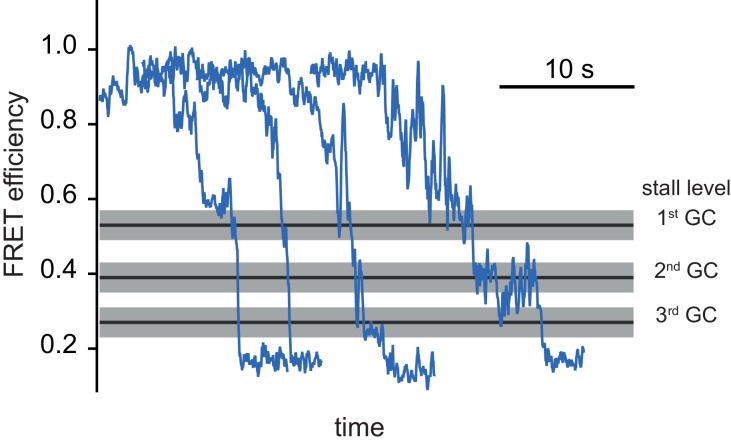
(a) Schematic illustration of the smFRET unwinding assay. After loading to the substrate G40P unwinds the dsDNA (containing between 0 and 3 consecutive GC base pairs after 10 AT base pairs) and thereby gradually separates the donor fluorophore from the acceptor fluorophore. Complete unwinding results in donor strand leaving from the surface. (b) Typical unwinding trace of G40P. At tinject the protein solution containing Mg.ATP is injected. The unwinding initiation time tinit depends on the protein concentration and is measured from the moment of protein solution injection until unwinding starts. The unwinding time tunw is determined from the moment of FRET efficiency decrease until donor strand leaving. (c) Typical FRET efficiency unwinding traces of G40P on an all AT substrate base pair. The scale bar indicates 1 s. (d) Typical FRET efficiency unwinding traces of G40P with one GC base pair. The helicase briefly stalls at a characteristic FRET efficiency or slips after stalling and unwinds in a second attempt. (e) Typical FRET efficiency unwinding traces of G40P on the substrate with two consecutive GC base pairs. The helicase stalls eventually at two distinct FRET efficiencies (see arrows). (f) Typical FRET efficiency unwinding traces of G40P on the substrate with three consecutive GC base pairs. The helicase rarely unwinds three consecutive GC base pairs completely (top trace). Most traces show unwinding attempts and stalls at distinct FRET efficiency levels (bottom trace). (g-i) FRET efficiency distribution of stall levels of the one GC (blue), two GC (black) and three GC base pair (red) substrate. A (multipeak) fit with Gaussians revealed one peak at EFRET = 0.52 ± 0.04 for 1GC, two peaks at EFRET = 0.53 ± 0.04 and at EFRET = 0.39 ± 0.04 (two dashed lines) for 2GC and three peaks at EFRET = 0.52 ± 0.04, EFRET = 0.38 ± 0.04 and at EFRET = 0.27 ± 0.04 (three dashed lines) for 3GC. (j) Average FRET efficiencies with stalls after 4AT, 7AT, 10AT and 13AT base pairs (black symbols) in comparison with peak positions of stalls induced by 1GC, 2GC and 3GC base pairs (blue symbols).