Fig. 2.
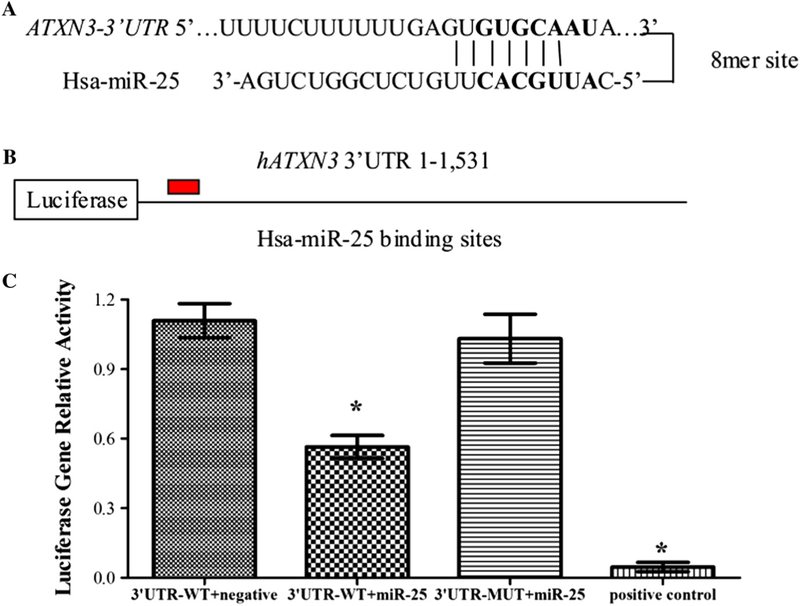
miR-25 directly targets the 3’ UTR of ATXN3. (A) The bioinformatics algorithms, including miRBase, PicTar, TargetScan and miRanda, identified the ATXN3 mRNA contains a miR-25 binding site at position 259–266 of the ATXN3 3’UTR. (B) The ATXN3 3’ UTR-WT (the 259–266 sequence was GTGCAATA) or ATXN3 3’ UTR-MUT (the 259–266 sequence was mutated to CTCGATAA) was fused to the 30 end of a luciferase gene. (C) Dual luciferase activity assays of co-transfections were performed using ATXN3 30 UTR-WT (with a partly miR-25 binding site), ATXN3 3’ UTR-MUT (with a mutated miR-25 binding site) or positive control (with a perfect miR-25 binding site) and miR-25 mimics. The results revealed that miR-25 mimics significantly reduced the expression of firefly luciferase when fused to the ATXN3 3’UTR-WT or positive control, but not the ATXN3 3’UTR-MUT. Error bars represent S.E.M.*P < 0.05 compared to the negative mimic control.
