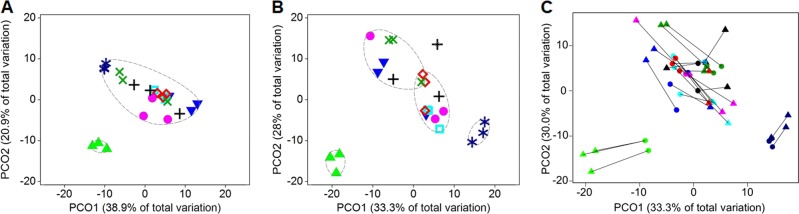
Fig. 3.
Community dissimilarity assessed by principal coordinates analysis (PCO) plots for all disturbance levels on T-RFLP datasets on days a 14 and b 35 of the study. Ovals with dashed lines represent 80% similarity calculated by group average clustering. Disturbance levels: L0 [light-green triangles], L1 [blue upside-down triangles], L2 [light-blue open squares], L3 [open red rhombuses], L4 [purple circles], L5 [black crosses], L6 [green x-symbols], and L7 [blue stars]. c Procrustes analysis on PCO at day 35 comparing metagenomics (circles) and T-RFLP (triangles) datasets. Lines unite data points from the same reactor (n = 24). Same colour palette as for disturbance levels. Tests comparing both methods were statistically significant (Supplementary Table 2). Intermediate treatments’ (L1–6) within-treatment dissimilarity increased with time. L0 and L7 clusters consistently displayed higher similarity after 14 days