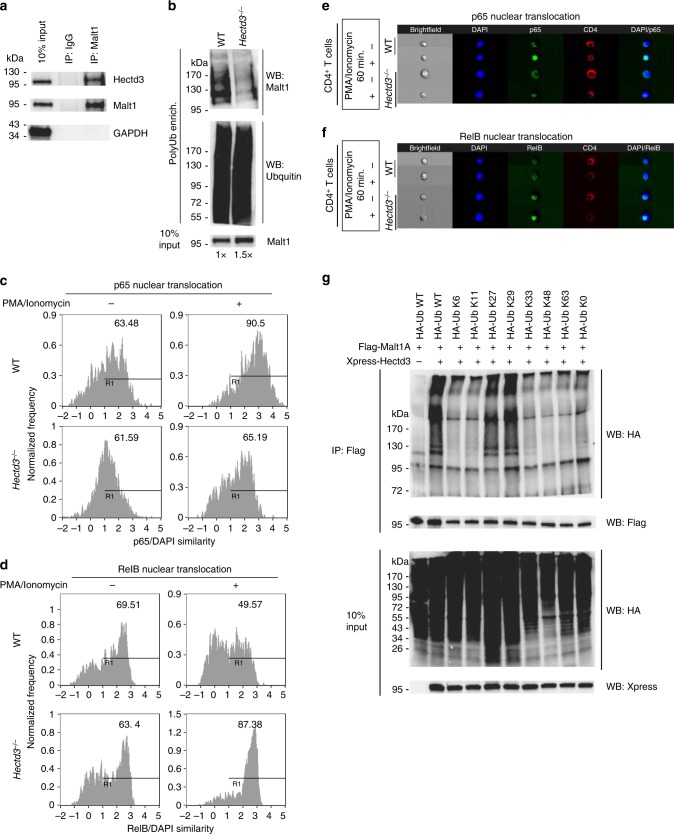
Fig. 5.
Hectd3 interacts and promotes non-degradative K27- and K29-linked polyubiquitination on Malt1 and NF-κB activation in CD4+ T cells. a Immunoblot of Hectd3, Malt1, and GAPDH following Malt1 or IgG immunoprecipitation of protein extracts from draining lymph node CD4+ T cells of WT mice 13 days following EAE induction. b Immunoblot of Malt1 following polyubiquitinated protein enrichment of extract from draining lymph node CD4+ T cells of wild-type Hectd3−/− or (WT) mice 13 days following EAE induction. About 1.5× the amount of total protein from Hectd3−/− CD4+ T cells, compared to WT CD4+ T cells, was used to normalize for the reduction in Malt1 protein level in Hectd3−/− CD4+ T cells. At least 700 μg of total protein from mouse primary CD4+ T cells were enriched for ubiquitinated protein with Ubiquitinated Protein Enrichment Kit or Anti-Ub TUBE2, Agarose following manufacturer’s protocol. c, d Percentage of p65 or RelB nuclear translocation using p65/DAPI similarity analysis from ImageStream, in CD4+ T cells isolated from draining lymph nodes of Hectd3−/− and WT mice, 13 days following EAE induction. e, f ImageStream fluorescence imaging of PMA/ionomycin-induced p65 nuclear translocation (e) or RelB (f) in CD4+ T cells isolated from draining lymph nodes of Hectd3−/− or WT mice, 13 days following EAE induction. g Immunoblot of HA, Flag, and Xpress following two-step Flag immunoprecipitation of protein extracts from HEK293T cells co-transfected with the indicated HA-Ub K only mutants, Flag-Malt1A, and Xpress-Hectd3. HA-Ub K only mutant denote that the only lysine in the HA-tagged ubiquitin is at the indicated residue, and all other lysine residues are mutated to arginine. HA-Ub K0 mutant indicate that all seven lysine residues are mutated to arginine. a–g Immunoblots, ImageStream similarity analyses, and ImageStream fluorescence imaging are representative of at least three independent experiments. Source data are provided as a Source Data file