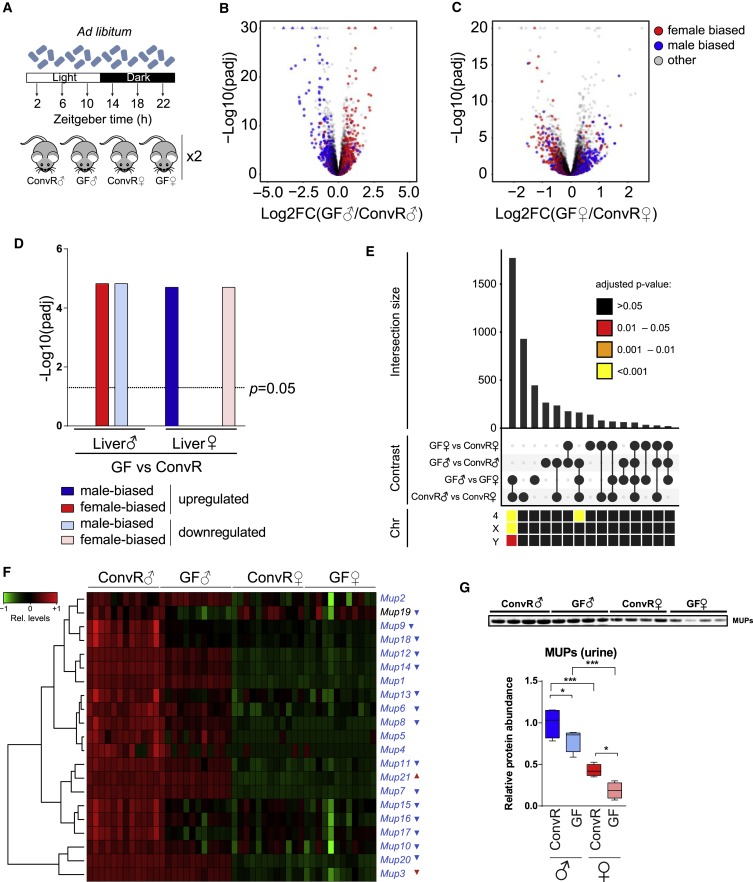
Figure 2.
A Lack of Gut Microbiota Decreases Sex-Specific Gene Expression by Feminizing Male and Masculinizing Female Hepatic Gene Expression
(A) Experimental design to detect sex-specific changes.
(B) Volcano plot of differences in mRNA expression between ConvR♂ and GF♂ categorized by sex-biased expression in ConvR mice (ConvR♂ versus ConvR female [ConvR♀]). Colors indicate male-biased (blue), female-biased (red), and unbiased (gray) expressed genes.
(C) Volcano plot of expression difference in mRNA expression between female GF (GF♀) and ConvR♀ mice. Color code is identical to (B).
(D) Statistical analysis of male- and female-biased genes showing a significant inversion of sex-biased genes toward the opposite sex in GF mice.
(E) Number of the shared differentially expressed genes (intersections size) between the four indicated contrasts. Lower heatmap indicates the enrichment for chromosomal location (Chr) for the corresponding intersection.
(F) Expression changes of differentially expressed Mup genes in GF♂ mice as assessed by RNA-seq. Most Mup genes are downregulated in GF♂ mice. The highest expressed Mup in female liver (Mup3) is downregulated in GF♀ mice. Colored gene name indicates unbiased (black) and male-biased (blue) expression. Triangle indicates the direction of significant change in GF♂ (blue) and GF♀ (red) mice in comparison with ConvR.
(G) Protein levels of MUP in the urine of GF♂ and GF♀ mice are significantly downregulated in male and female mice compared with their ConvR counterparts (each Tukey boxplot represents four independent biological replicates per condition). ∗p ≤ 0.05, ∗∗∗p ≤ 0.001.