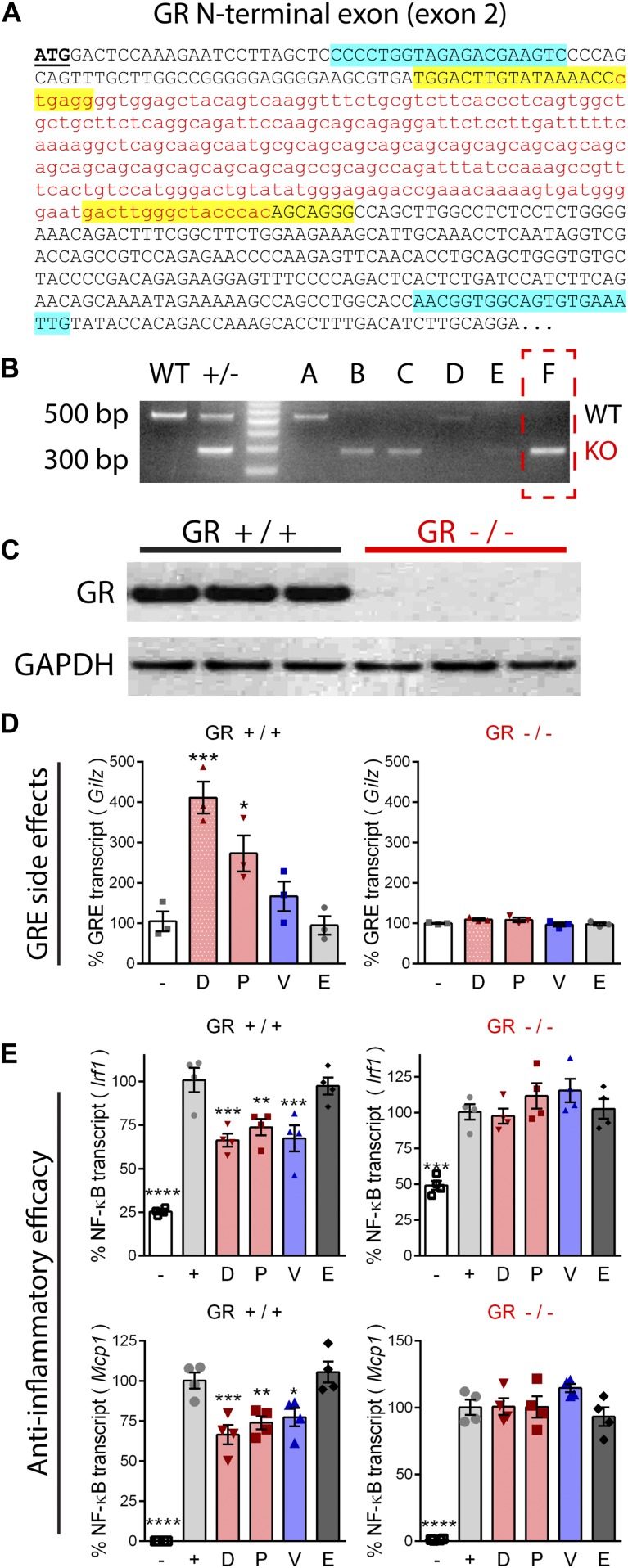
Figure 2. CRISPR knockout of the GR shows that anti-inflammatory pathways are GR-dependent.
A frame-shifting deletion mutation was introduced via CRISPR in C2C12 myoblasts to knockout GR expression. (A) The DNA sequence of the GR N-terminus is provided. Base pairs deleted in knockout cells are shown as red, lower case letters. Positions of CRISPR guide RNAs are highlighted in yellow, and genotyping primers in blue. (B) PCR of C2C12 myoblast clones (labeled A through F) identified WT (∼500 bp band) and GR deletion clones (∼300 bp band). (C) Western blot confirmed GR knockout in clone from PCR lane F. (D) Drug-treated wild-type and GR knockout myoblasts were assayed for activation of a GR transactivation target transcript (Gilz). (E) Wild-type and GR knockout myotubes were treated with drug, induced with TNF, and assayed for expression of inflammatory genes regulated by NF-κB (Irf1 and Mcp1). (n = 3–4, *P < 0.05, **P < 0.005, ***P < 0.001, ****P < 0.0001, ANOVA with post hoc versus vehicle [panel D] or + TNF [panel E] control; representative of three experiments; panels [D, E]: [−] = no drug or TNF, [+] = TNF plus vehicle, D = deflazacort, P = prednisolone, V = vamorolone, E = eplerenone).