Figure 1.
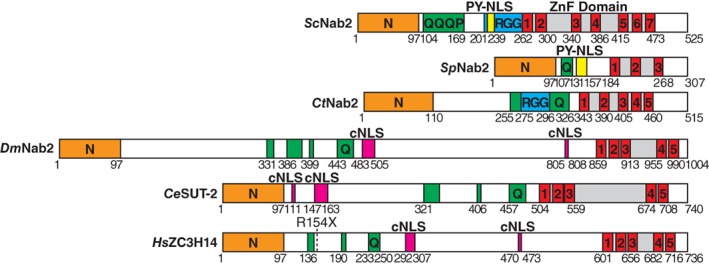
Members of the Nab2/ZC3H14 zinc finger polyadenosine RNA binding protein family have a similar domain architecture. Nab2 orthologues – S. cerevisiae Nab2 (ScNab2; Uniprot ID: P32505), S. pombe Nab2 (SpNab2; Uniprot ID: O13713), C. thermotolerans Nab2 (CtNab2; Uniprot ID: G0SCL7), Drosophila melanogaster Nab2 (DmNab2; Uniprot ID: Q9V3H9), Caenorhabditis elegans SUT‐2 (CeSUT‐2; Uniprot ID: Q95XU6), and Homo sapiens (HsZC3H14; Uniprot ID: Q6PJT7) share a common domain structure based on a N‐terminal PWI‐fold domain (orange), which serves as a protein–protein interaction domain in ScNab2, followed by a Q‐rich region (green), and a C‐terminal Zn finger (ZnF) domain (gray) containing a series of Zn fingers (red), which bind to polyadenosine RNA. ScNab2 and CtNab2 contain an RGG domain (blue), which functions in karyopherin‐based nuclear import in ScNab2. In addition, ScNab2 contains a Pro‐Tyr nuclear localization signal (PY‐NLS) (yellow) and SpNab2 contains a predicted PY‐NLS that functions in nuclear import in ScNab2. Nab2 orthologues from higher Eukaryotes contain two predicted classical nuclear localization signals (cNLS) (magenta) that function in karyopherin‐based nuclear import. The nonsense mutation R154X identified in ZC3H14 in individuals with autosomal recessive intellectual disability is highlighted.
