Figure 3.
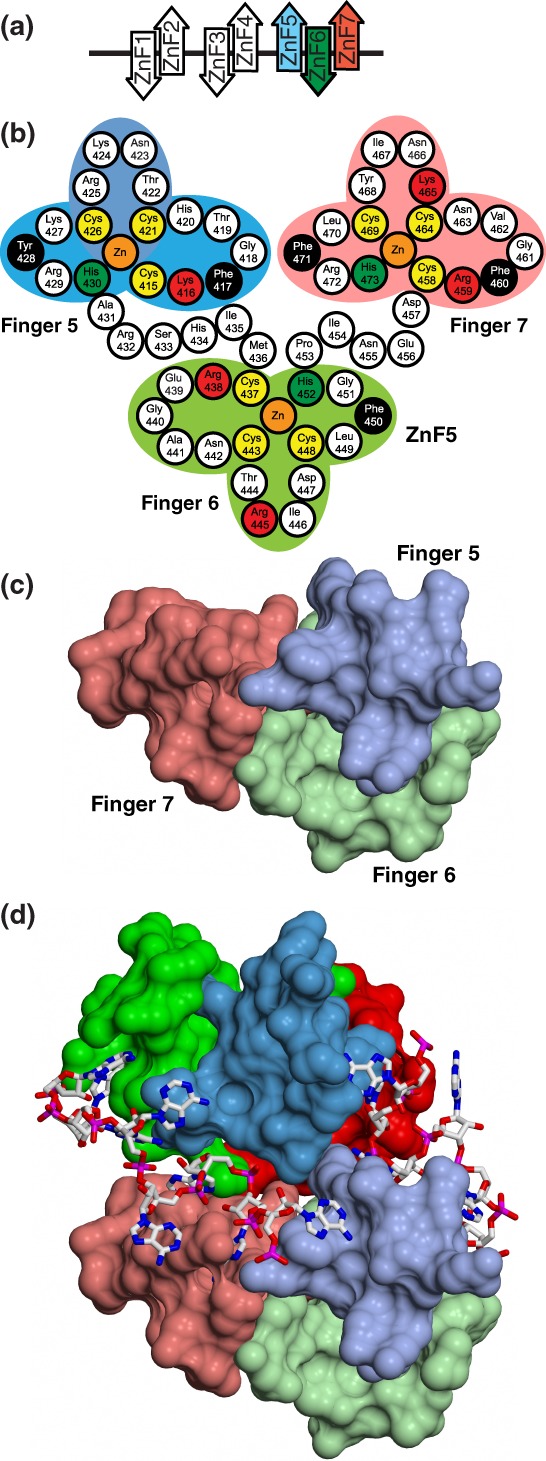
Nab2 Zn fingers. (a) Schematic illustration of the structures of the S. cerevisiae Nab2 Zn fingers determined by NMR (28, 43) and X‐ray crystallography.29, 30 The Zn fingers are clustered into three groups, and the fingers within each group have a defined orientation that influences their interaction with adenine‐rich RNA chains. (b) Arrangement of residues in ScNab2 Zn fingers 5–7. Key hydrophobic and basic residues that are important for binding to adenines (see also Fig. 4) are black and red, respectively. (c) Structure of ScNab2 Zn fingers 567. Zn finger 6 (green) projects on the opposite of the module to Zn fingers 5 (blue) and 7 (red). (d) Heterotetramer produced by A11G binding to ScNab2 Zn fingers 567 in which each RNA chain binds to both protein chains. Based on PDB 5L2L Panel (B) is derived from C. Brockmann et al. (2012) “Structural basis for polyadenosine‐RNA binding by Nab2 Zn fingers and its function in mRNA nuclear export.” Structure, 20:1007–1018.
