Abstract
The Patient Health Questionnaire–9 (PHQ-9) is a validated instrument for assessing depression severity. While some electronic health record (EHR) systems capture PHQ-9 scores in a structured format, unstructured clinical notes remain the only source in many settings, which presents data retrieval challenges for research and clinical decision support. To address this gap, we extended the open-source Leo natural language processing (NLP) platform to extract PHQ-9 scores from clinical notes and evaluated performance using EHR data for n=123,703 patients who were prescribed antidepressants. Compared to a reference standard, the NLP method exhibited high accuracy (97%), sensitivity (98%), precision (97%), and F-score (97%). Furthermore, of patients with PHQ-9 scores identified by the NLP method, 31% (n=498) had at least one PHQ-9 score clinically indicative of major depressive disorder (MDD), but lacked a structured ICD-9/10 diagnosis code for MDD. This NLP technique may facilitate accurate identification and stratification of patients with depression.
Introduction
Depression is one of the leading causes of disability and a major contributor to the overall global burden of disease, according to World Health Organization.1 The National Institute of Mental Health estimated that 16.2 million adults in the United States had a major depressive episode in 2016.2 Left untreated, depression increases the risk for morbidity, suicide, decreased cognitive and social functioning, self-neglect, and early death.3 While depression can be treated, it often goes undiagnosed.4 Although primary care is an ideal setting to identify and offer treatment for depression, in nearly 50% of cases physicians fail to diagnose major depressive syndromes in their patients.5 New national guidelines recommend that depression should be screened and further evaluated before initiating treatment.6
Approaches for clinicians to document depression severity include the Patient Health Questionnaire-9 (PHQ-9) and the International Classification of Disease Ninth Revision (ICD-9) and Tenth Revision (ICD-10) diagnosis codes. The PHQ-9 is a validated self-report questionnaire pertaining to the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) criteria of major depression.7 An overall PHQ-9 score ranges from 0 to 27, and scores of 10 or above are highly associated with major depressive disorder (MDD).8 In addition to PHQ-9 scores, clinicians often document ICD-9 and ICD-10 diagnosis codes mainly used for billing. However, studies have demonstrated limitations of diagnosis codes for patient cohort identification.9
Electronic health records (EHR) are emerging as a major source of data for clinical and translational research.10 Most EHR systems capture PHQ-9 score only in unstructured clinical notes such as encounter and psychiatry notes, which presents challenges for automated extraction to support research and clinical decision support applications. In contrast, structured data, such as ICD-9 and ICD-10 diagnosis codes, are more amenable to structured queries, but are of limited utility for accurately identifying clinical phenomena of interest. Several studies have demonstrated the value of natural language processing (NLP) methods applied to unstructured notes in a number of clinical areas, including asthma11 detection, pneumonia detection12, colonoscopy testing13, and cervical cancer screening.14
To the best of our knowledge, very few studies have investigated NLP for patients with depression.15,16 These studies have used sophisticated machine learning methods and expert-defined lists of terms to classify depression status in unstructured notes. Of note, the classifiers described in these studies did not use PHQ-9 scores, although Perlis and colleagues identified PHQ-9 scores as valuable for future work to improve classifier performance.15 Additionally, the existing studies of NLP for depression used different NLP systems, which can limit portability of NLP methods across settings.17,18
Previously our group has demonstrated the benefit of approaching “low-hanging NLP tasks” that exploit note metadata, select patterns with low sensitivity with respect to location in a note, adapt simple rule-based extraction logic, and use existing NLP software to encourage portability. In contrast, “hard-to-reach NLP tasks” often involve development of heuristics, knowledge engineering, machine learning, and novel systems. PHQ-9 scores in unstructured notes represent under-utilized measures of depression severity, and their extraction may represent a “low-hanging NLP task” that can inform other efforts in depression research and practice.15 In this study, our goal was to extend the open-source Leo NLP platform19, developed by the Veteran’s Administration, to extract PHQ-9 scores from clinical notes and evaluate performance using data from our institution.
Materials and Methods
Setting
Weill Cornell Medicine (WCM), an academic medical center in New York City on the Upper East Side of Manhattan, hosts approximately 1000 physicians, who work at over 20 outpatient sites across the metropolitan area to treat hundreds of thousands of patients annually. WCM physicians hold admitting privileges for patients at NewYork-Presbyterian Hospital. In the outpatient setting, WCM physicians use the EpicCare® Ambulatory EHR platform to document clinical care, from which we extracted the various clinical notes used as the basis of this study. This study was approved by the WCM Institutional Review Board (IRB).
Data collection
In order to develop and validate our NLP method for extracting PHQ-9 scores, we obtained all clinical notes for 123,703 patients who were prescribed an antidepressant, as defined by the Healthcare Effectiveness Data and Information Set (HEDIS).20 The EHR data contained over 13.8 million clinical notes for this cohort, authored between 2007 and 2017. Clinical notes that we used for the performance evaluation and data analysis were authored by clinicians from multiple specialties (e.g., internal medicine, psychiatry, anesthesiology, pain medicine) in multiple office locations. Notes were highly heterogeneous in their content and level of details and unstructured in their format. Instances of the PHQ-9 concept in these notes often occurred in short snippets with telegraphic structure or unusual punctuation. Table 1 shows several examples of snippets of PHQ-9 concepts—often with an associated score—that appear in these notes, illustrating the lexical variation with which the PHQ-9 concept is described in this corpus.
Table 1.
Examples of PHQ-9 instances expressed in clinical free text notes.
| Text snippets |
|---|
| Depression: PHQ-9 score 16 6/14 |
| positive PHQ9 |
| refusing phq 9 |
| PHQ-9: 19/27 score |
| PHQ 2/27 |
| PHQ-9 score 13 (not disabled) |
| PHQ-9 score today 11 |
| PHQ-9 score – 5 |
| PHQ9 > 5 |
| PHQ 9--> 9 with some SI |
| 17 on PHQ 9 |
| PHQ-9 16/28; |
| PHQ indicates moderate depression |
| PHQ-9 18/27 |
| PHQ-9 = 5 |
| PHQ=15/27 |
| PHQ 23/27 but no suicidal intent although sometimes she feels that she would be better off dead |
| Depression: PHQ-9 21 |
Reference standard creation
We developed the reference standard through manual review of 1,000 encounter notes selected at random from a superset of notes containing the character string “phq”. To establish the reference standard, two reviewers (PA, JD) annotated all notes based on pre-defined guidelines. These guidelines included reading all notes to identify all mentions of the PHQ-9 concept and any associated quantitative score. We observed 98 percent agreement between the two reviewers. In 19 cases, the two reviewers differed in their assessment: for these cases, a third reviewer (ES) serving as adjudicator resolved the discrepancy. The reviewers also confirmed all notes that did not have any mention of PHQ-9 information.
For each document in the dataset, the reviewer identified a numeric value between the range 0 and 27 for PHQ-9. In notes that contained multiple instances of PHQ-9, we took the last occurrence of PHQ-9 and the corresponding value pair to represent that document. Similarly, some notes mentioned PHQ-9 simply by the term “PHQ” without explicitly mentioning the nine-item questionnaire. If the value associated with such mention is expressed on a reference base of 27 (e.g. 6/27), then we treat it as PHQ-9 concept. Also in some reports, the PHQ-9 value is expressed using a greater than or less than symbol (e.g. PHQ-9 > 5). In these cases the reviewer extracted the value ignoring the symbol. In reports where there was no quantitative value for PHQ-9 available, we made no attempt to assign a numerical value, even though depression severity is mentioned in qualitative terms such as ‘mildly’, ‘moderately’ or ‘severely’ depressed.
Leo NLP system
The Leo architecture is a collection of services and libraries that facilitate the rapid creation and deployment of annotators for NLP using the Apache Unstructured Information Management Architecture - Asynchronous Scaleout (UIMA-AS).19,21,22 Leo consists of three main components: client, core, and service. The client handles data input and output, while the core component contains tools for NLP annotation (developed locally or acquired from elsewhere). UIMA-AS provides capabilities for efficient, real-time processing and can scale up to handle millions of clinical text documents. The service component contains UIMA-AS services, which define a type system and annotators in a pipeline to implement logic for extracting specific concepts from unstructured notes. Leo services can be deployed in multiple ways, the simplest being a single instance of a pipeline executed in a synchronous manner. More complex configurations may host multiple services with multiple instances of a pipeline, and can be distributed across multiple network servers executed in an asynchronous manner. Installation of Leo involves downloading and extracting the content of the distribution package, setting up environmental variables, and configuring settings for the Leo reader and listener services.
NLP method development
To extract PHQ-9 scores from clinical notes, a new instance of Leo, named PhqExtractor was developed and implemented. Previously, we demonstrated the portability of Leo from the Veterans Administration to our institution for extraction of ejection fraction values from unstructured notes.18 We employed a similar approach to create an annotation pipeline to extract PHQ-9 scores from unstructured notes.
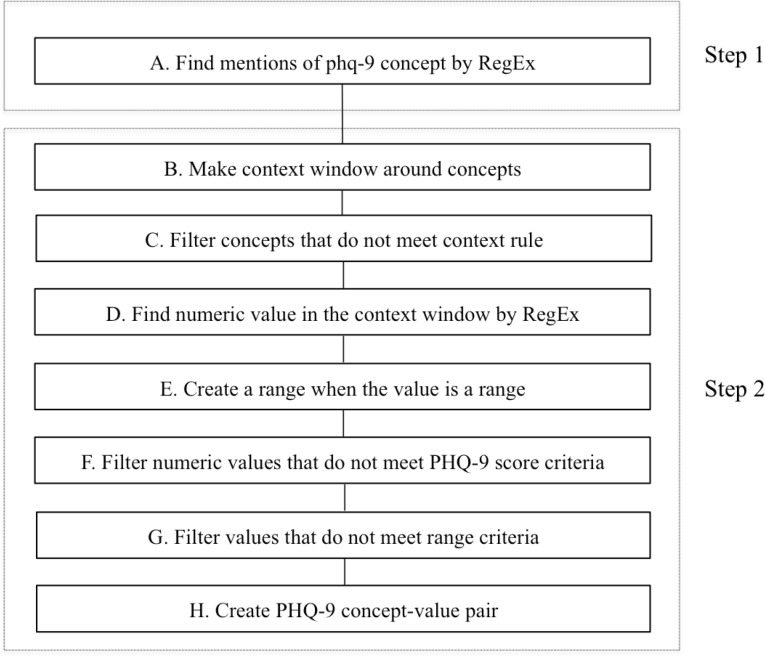
The creation of extraction logic is an iterative two-step process that includes concept definition, context analysis, rule definition, system application, and error analysis (Figure 1). Step 1 of development involves identifying the set of core concepts for PHQ-9. Regular expressions, string matching, and filters were used to extract the concepts. Concepts were identified using multiple regular expressions that allowed for variation in expression. Patterns for keywords such as “PHQ” and “patient health questionnaire” were defined. In step 2, iterative context analysis was used to determine if these keywords were mentioned in the context of the nine-item PHQ-9 instrument. A window of appropriate surrounding words was then defined for specific rule generation. Three types of rules were developed: a) positive context rules defining phrases that indicate the relevant presence of the concept of interest; b) non-positive context rules defining phrases that indicate that the concept of interest is not present or undetermined; c) exclusion context rules defining terms that are similar to the target concepts. We excluded instances of other PHQ measures, such as PHQ-2, PHQ-4 and PHQ-7.
Figure 1.
NLP pipeline logic implemented for extracting PHQ-9 scores from clinical notes
Quantitative values for PHQ-9 were found using number patterns, allowing for with or without modifiers such as ‘=,’ ‘(,’ ‘>,’ ‘(<,’ and ranges of values. Since each item in PHQ-9 is rated on a 4-point scale from 0 to 3 and the overall scores can range from 0 to 27, only numeric values in this range were considered as proper scores. Some instances of PHQ-9 are mentioned only by the “PHQ” term and disambiguation of such a term from other PHQ instruments such as PHQ-2 or PHQ-7 is problematic. To eliminate false detection of non-PHQ-9 PHQ values, our extraction logic treated such instances as PHQ-9 only in instances where the associated value was mentioned with relation to a base score of 27, e.g. “PHQ 18/27”. This improves the overall detection of true positive cases of PHQ-9 instances. Similarly, in some notes, clinicians expressed PHQ-9 scores in a long question-answer list format containing the 9 individual items and their corresponding scores on the PHQ-9 questionnaire. The cumulative score mentioned for such instances falls outside the window that was defined for various rule generations within the initial extraction logic. A custom annotation type and additional rules were implemented to extract scores for such PHQ-9 instances. For the rule-based algorithm development and testing, we used 1000 randomly selected notes from a subset of notes previously identified by SQL query as having a mention of a character string of “phq”. Although this approach may have the potential to introduce some input bias, our main rationale was to increase the number of positive cases of PHQ-9 in our NLP algorithm development since less than one percentage of the larger set have a “phq” mention in them. The development set was further divided into four batches comprised of 100, 200, 300, and 400 reports. We used an initial set of regular expressions and rules for PHQ-9 to develop the system for the first batch of 100 reports. Successive batches of development subsets were then used to refine the regular expressions and rules to maximize the accuracy. We identified instances where existing rules failed to extract information and therefore made further modification to the pattern sets. These steps were repeated, updating the pattern iteratively until the system extracted all possible instances of PHQ-9.
For each note, the NLP method attempted to extract all PHQ-9 instances and the associated scores. Therefore, in the post-processing stage, we used a custom mapping tool to associate a given instance of PHQ-9 for each note. The logic behind this mapping follows the same heuristic used for creating the reference standard. In notes where there is only one PHQ-9 score extracted, we associated that score with the note. However, when the NLP method extracted multiple PHQ-9 scores from one note, we associated only the last-mentioned PHQ-9 score with that note.
Evaluation of the NLP method
To evaluate the NLP method, we used the reviewer-annotated reference standard data set consisting of 1,000 notes as input into the Leo platform. The output of the NLP method was tabulated and compared against the reference standard annotations. Each note was classified as one of four possible cases.
A true positive was defined as an instance where the PhqExtractor pipeline successfully extracted the PHQ-9 score defined in the reference standard.
A false positive was defined as an instance where the PhqExtractor pipeline extracted a PHQ-9 value but the reference standard did not contain a PHQ-9 value.
A true negative was defined as an instance where the PhqExtractor pipeline did not extract a PHQ-9 value and the reference standard did not contain a PHQ-9 value.
Finally, a false negative was defined as either an instance where the PhqExtractor pipeline did not extract a PHQ-9 value but the reference standard contained a PHQ-9 value, or an instance where the PhqExtractor pipeline extracted a PHQ-9 value that did not match the reference standard.
We then used counts of the four cases to construct a confusion matrix and calculate precision, accuracy, recall, and F-score.
After evaluating performance of the NLP method on 1,000 notes (675 unique patients) comprising the reference standard, we then applied the NLP method to extract PHQ-9 scores from all notes in the study cohort containing a mention of “phq” as per SQL query. We determined how many patients had at least one PHQ-9 score documented and how many patients lacked documentation of a PHQ-9 score.
Utilizing SQL queries against outpatient EHR data in our data warehouse23, we then determined the extent to which patients in the study cohort had structured ICD-9 and/or ICD-10 diagnosis codes for depression and MDD documented. For defining depression, we identified interface terminology items corresponding to ICD-9 codes starting with 311.*, 300.4*, 292.2*, or 292.3* (equivalent to ICD-10 codes F32.* and F34.1*). Similarly, for defining major depressive disorder, we identified interface terminology items corresponding to ICD-9 codes starting with 292.2 or 292.3 (equivalent to ICD-10 codes F32.*). Interface terminology integrated with our instance of Epic allows for queries referencing ICD-9 codes to also return instances of diagnoses associated with ICD-10 codes utilizing the many-to-many mapping intrinsic to the interface terminology. Sources of diagnosis codes in the EHR included the patient’s problem list, medical history, encounter diagnoses, and billing diagnoses. For patients with at least one note containing a PHQ-9 score, we then determined how many patients had a PHQ-9 score greater than 10, a threshold for defining MDD8. We then determined how many patients in each group had an ICD-9 or ICD-10 code for MDD.
Results
For extracting PHQ-9 scores from the 1,000 notes comprising the reference standard, the NLP method yielded 742 true positives, 217 true negatives, 25 false positives, and 16 false negatives. These values resulted an overall accuracy of 96% (95% CI 94% - 97%), sensitivity of 98% (95% CI 97% - 99%), specificity of 90 (95% CI 87% - 92%), positive predictive value 97% (95% CI 96% - 98%), negative predictive value of 93 (95% CI 90% - 96%), and an F-score of 97%. When applying the NLP method to all notes in the cohort, we identified 1,583 patients who had at least one note containing a PHQ-9 score. It should be noted that this is only 1.2 percentage of our patient cohort. We evaluated on a testing set known to contain the string value of “phq” – a relatively small subset of over 11.7 million documents. It is unlikely that the good precision and recall from the testing set would extend to the broader set. Table 2 describes characteristics of patients that comprised the reference standard, patients that had any PHQ-9 scores extracted, and patients that did not have any PHQ-9 scores extracted.
Table 2.
Characteristics of patient’s demographic investigated in this study.
| Patients in reference standard (n=675) | Patients that had any PHQ-9 scores extracted (n=1,583) | Patients that did not have any PHQ-9 scores extracted (n=122,120) | |
| Age, mean years(±SD) | 54 (±19.42) | 53 (±19.05) | 56 (±18.01) |
| Gender, n (%) | |||
| Female | 442 (65%) | 1,037 (66%) | 81,237 (66%) |
| Male | 233 (35%) | 546 (34%) | 40,865 (33%) |
| Unknown | 0 | 0 | 18 (<1%) |
| Ethnicity, n (%) | |||
| Hispanic | 100 (14%) | 187 (12%) | 7649 (6%) |
| Non-Hispanic | 344 (51%) | 845 (53%) | 48,346 (40%) |
| Declined/Unknown | 231 (34%) | 551 (35%) | 66,125 (54%) |
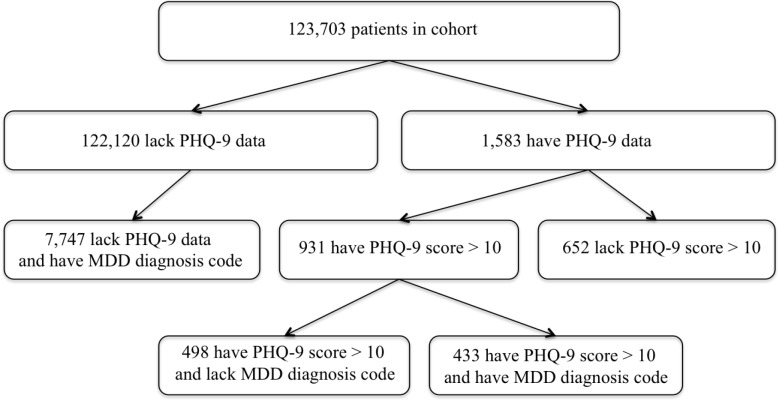
As shown in Figure 2, of the 1,583 patients with a PHQ-9 score extracted by the NLP method, 931 (58%) had a PHQ-9 value greater than 10, a common threshold for defining MDD. Of these 931 patients, 498 (53.4%) never received a structured diagnosis of MDD, and 51 (5.5%) never received a structured diagnosis of any form of depression. The majority of PHQ-9 values greater than 10 in these patients were assigned at visits with providers specializing in geriatric and internal medicine.
Figure 2.
Extent of structured diagnosis data for depression in patients with NLP-derived PHQ-9 scores
The source code and documentation for the NLP method are available for download at https://github.com/wcmc-research-informatics/PhqExtractor.
Discussion
This study demonstrated the effectiveness of approaching a “low-hanging fruit” NLP task: the extraction of PHQ-9 scores from unstructured clinical notes. To our knowledge, no other studies have targeted this concept for NLP-based extraction. The utility of this NLP technique lies principally in its potential to facilitate greater accuracy and precision in identifying patients with depression and further distinguishing between varying grades of depression severity. Through our PhqExtractor technique, we were able to determine that of the 1,583 patients with an extracted PHQ-9 value greater than 10, the majority had never received a structured diagnosis consistent with major depressive disorder. The extraction logic we have demonstrated here for PHQ-9 values could be easily modified to extract other mental health concepts, such as Generalized Anxiety Disorder (GAD) measures and Clinical Global Impression scale (CGI), from unstructured notes. We should caution here that although PhqExtractor performed very well as suggested by the high sensitivity (recall) and positive predictive value (precision) when applied to a subset set of notes that contains the term “phq”, it is unlikely that these performance would extend to a broader set. We evaluated the testing set but not the broader set notes. We assume the precision and recall generalize.
The well-documented association between PHQ-9 scores greater than 10 and major depressive disorder suggests that at least some of these patients may suffer from MDD. Evidence suggests that clinical diagnostic workflows are often driven by ease-of-use concerns24: many of the patients we observed with high PHQ-9 scores but no diagnosis of major depressive disorder had received diagnoses of “Depression, unspecified” (corresponding to an ICD-9 code of 311.*), suggesting that physicians were choosing the option that required the least amount of typing and the fewest number of clicks. Of the providers who did not assign diagnoses of MDD to patients with PHQ-9 scores greater than 10, the majority specialized in internal medicine and geriatric medicine – psychiatrists were more likely to assign MDD diagnoses to patients with high PHQ-9 scores. Future work may assess the extent to which the differing extent of structured diagnostic documentation may extend to the PHQ-9 scores themselves, examining whether psychiatrists were more or less likely to assign higher scores. Additionally, concern with social stigma related to depression, as well as patient concerns regarding the presence of a formal diagnosis of depression, may lead physicians to avoid entering a structured diagnosis code in the patient’s EHR. It may be possible to increase the accuracy of existing structured diagnosis documentation within the EHR using clinical decision support modules within the electronic health record that prompt physicians to assign a diagnosis of major depressive disorder when recording a PHQ-9 value greater than 10.
Scores on PHQ-9 and similar instruments are just one of many concepts and terms in narrative notes that physicians use in ascertaining depression severity9. Direct mentions of concepts such as “severe depression”, “suicidal ideation”, “suicidal thoughts” etc., are indicative of depression in varying degrees, and proper retrieval and interpretation of such concepts requires more complex extraction logic. Although it may be possible to capture the timeline for PHQ-9 scores recorded for a specific patient, we made no attempt to distinguish between acute and chronic depression in patients over time, seeking rather to establish a reliable methodology for extraction of PHQ-9 values first. Future research may focus on longitudinal assessment of a patient’s progression through the course of remission and relapse. This study analyzed clinical notes of those patients who were prescribed a given class of antidepressant medications. Vast majority of these notes, however, do not have mentions of PHQ-9. It would be interesting to compare PHQ-9 scores extracting from narrative notes originating from multiple specialties.
The construction of computable phenotypes has been well documented as an approach towards leveraging EHR data to facilitate both cohort identification and secondary use of structured data both within and between institutions.25-27 Integrating NLP-derived PHQ-9 scores with other structured data elements derived from the EHR offers the potential to enable the identification of patients suffering from depression or depressive symptoms with greater specificity and accuracy compared to simple code-based approaches. Physicians seeking to recruit patients for clinical trials or prospective research in studying depression may find that they are unable to identify a sufficient number of patients as determined by power analysis using ICD-9/10 codes alone, and that supplementing with PHQ-9 values serves as a valuable additional source of potential participants. Adding the criteria of an NLP-derived PHQ-9 value greater than 10 to a trial’s inclusion criteria (in addition to a formal structured diagnosis of major depressive disorder) at our institution would lead to an increase of about 5% to the size of the patient cohort. Alternately, researchers may find that ICD-9/10 codes lack sufficient positive predictive value to accurately identify subjects, and that PHQ-9 values allow them to better identify potential participants.
While NLP systems hold great potential in patient care and clinical research, portability of these tools across multiple sites remains a challenge. Several strategies have been suggested to mitigate these challenges. We recently demonstrated a practical strategy for NLP portability using extraction of left ventricular ejection fraction (LVEF) as a use case.18 This study is another example of such an approach, where a relatively simple, rule-based NLP solution proved to be very effective. Our experience in implementing this approach to extract PHQ-9 values further illustrates the point that simple NLP applications may be easier to disseminate and adapt, and in the short term may prove more useful, than complex applications which often require more advanced methods and may be difficult to port across sites.
Manual review of the notes classified as false positive and false negative suggests potential directions for further improvement to our extraction logic. One frequent confounder causing false positives was the close proximity of other clinical concepts, such as generalized anxiety disorder (GAD) or medication information, to the PHQ-9 concept. For example, in the following statements: “Did PHQ9 and GAD7 again today” or “Psych- Up zoloft to 100mg by 25 mg. Keep log by PHQ9” or “PHQ-9 Questionnaire- N/A HCV Dx: 2003,” our extraction logic wrongly identified the neighboring numeric value as the score for PHQ-9. During the development phase of rule generation we tested with varying window sizes on both sides of the PHQ-9 concept in order to minimize these false positive cases. However, the abbreviated format often used to document PHQ-9 scores made it difficult to find an appropriate window size without adversely impacting sensitivity by ruling out other valid instances of PHQ-9 values. The simple heuristic of selecting the last occurrence of a PHQ-9 instance as representative of a document also introduced some false negative classification. Similarly, the filtering rules we developed were not adequate to correctly identify all possible scenarios with which clinicians expressed PHQ-9 concepts in their notes. For example, in statements such as “Pt had been referred by PCP after scoring 13 on PHQ 9 (with a 1 on the question of thinking you would be better off dead).”, or “PHQ=8/27 GAD=10/21 10/20/15. PHQ 10/26/15 =5/27”, our filtering logic failed to extract the correct PHQ-9 score from multiple numerical values.
Limitations of this study include its conduct at a single site. However, code is freely available on GitHub to encourage adoption by researchers at other institutions, where clinical documentation practices may vary. The NLP system described in this study is built on an open source UIMA architecture that is freely accessible and therefore encourages generalizability. Additionally, due to IRB protocol restrictions, our study was limited to a specific cohort of patients prescribed antidepressants. Future research may include expansion of the evaluation of the PhqExtractor pipeline on a broader patient cohort, as well as testing performance on notes originating from multiple sites. We also plan to evaluate NLP techniques designed to identify depression and its related sequelae, including suicidal ideation and suicide attempts.
Depression is a major public health concern that has not, to date, received as much attention from the NLP research community as other disease areas. It is our hope that this study contributes towards closing this gap.
Acknowledgments
This study received support from NewYork-Presbyterian Hospital (NYPH), Weill Cornell Medical College (WCMC), including the Clinical and Translational Science Center (CTSC) (UL1 TR000457), Joint Clinical Trials Office (JCTO) and R01 MH105384. We thank Drs. Olga V. Patterson and Scott L. DuVall of VA Salt Lake City Health Care System for their support and technical assistance in implementing the Leo NLP system at Weill Cornell Medicine.
References
- 1.Ferrari AJ, Charlson FJ, Norman RE, Patten SB, Freedman G, Murray CJL, et al. Burden of depressive disorders by country, sex, age, and year: findings from the global burden of disease study 2010. PLoS Med. 2013 Nov 5;10(11):e1001547. doi: 10.1371/journal.pmed.1001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.NIMH » Major Depression [Internet] Major Depression. [cited 2018 Feb 21] Available from: https://www.nimh.nih.gov/health/statistics/major-depression.shtml.
- 3.Olfson M, Blanco C, Marcus SC. Treatment of adult depression in the united states. JAMA Intern Med. 2016 Oct 1;176(10):1482–91. doi: 10.1001/jamainternmed.2016.5057. [DOI] [PubMed] [Google Scholar]
- 4.Fiske A, Wetherell JL, Gatz M. Depression in older adults. Annu Rev Clin Psychol. 2009;5:363–89. doi: 10.1146/annurev.clinpsy.032408.153621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wittchen H-U, Mühlig S, Beesdo K. Mental disorders in primary care. Dialogues Clin Neurosci. 2003 Jun;5(2):115–28. doi: 10.31887/DCNS.2003.5.2/huwittchen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gautam S, Jain A, Gautam M, Vahia VN, Grover S. Clinical Practice Guidelines for the management of Depression. Indian J Psychiatry. 2017 Jan;59(Suppl 1):S34–50. doi: 10.4103/0019-5545.196973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manea L, Gilbody S, McMillan D. A diagnostic meta-analysis of the Patient Health Questionnaire-9 (PHQ-9) algorithm scoring method as a screen for depression. Gen Hosp Psychiatry. 2015 Feb;37(1):67–75. doi: 10.1016/j.genhosppsych.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 8.VA/DoD Clinical Practice Guidelines - Management of Major Depressive Disorder (MDD) [Internet] US Department of Veterance Affiars. [cited 2018 Feb 26]. Available from: https://www.healthquality.va.gov/guidelines/MH/mdd.
- 9.Wei W-Q, Teixeira PL, Mo H, Cronin RM, Warner JL, Denny JC. Combining billing codes, clinical notes, and medications from electronic health records provides superior phenotyping performance. J Am Med Inform Assoc. 2016 Apr;23(e1):e20–7. doi: 10.1093/jamia/ocv130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao KP, Cai T, Savova GK, Murphy SN, Karlson EW, Ananthakrishnan AN, et al. 2015. Apr 24, Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ; pp. 350–h1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu ST, Sohn S, Ravikumar KE, Wagholikar K, Jonnalagadda SR, Liu H, et al. Automated chart review for asthma cohort identification using natural language processing: an exploratory study. Ann Allergy Asthma Immunol. 2013 Nov;111(5):364–9. doi: 10.1016/j.anai.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones BE, South BR, Shao Y, Lu CC, Leng J, Sauer BC, et al. Development and Validation of a Natural Language Processing Tool to Identify Patients Treated for Pneumonia across VA Emergency Departments. Appl Clin Inform. 2018 Jan;9(1):122–8. doi: 10.1055/s-0038-1626725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denny JC, Peterson JF, Choma NN, Xu H, Miller RA, Bastarache L, et al. Extracting timing and status descriptors for colonoscopy testing from electronic medical records. J Am Med Inform Assoc. 2010 Aug;17(4):383–8. doi: 10.1136/jamia.2010.004804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagholikar KB, MacLaughlin KL, Henry MR, Greenes RA, Hankey RA, Liu H, et al. Clinical decision support with automated text processing for cervical cancer screening. J Am Med Inform Assoc. 2012 Oct;19(5):833–9. doi: 10.1136/amiajnl-2012-000820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perlis RH, Iosifescu DV, Castro VM, Murphy SN, Gainer VS, Minnier J, et al. Using electronic medical records to enable large-scale studies in psychiatry: treatment resistant depression as a model. Psychol Med. 2012 Jan;42(1):41–50. doi: 10.1017/S0033291711000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou L, Baughman AW, Lei VJ, Lai KH, Navathe AS, Chang F, et al. Identifying Patients with Depression Using Free-text Clinical Documents. Stud Health Technol Inform. 2015;216:629–33. [PubMed] [Google Scholar]
- 17.Carrell DS, Schoen RE, Leffler DA, Morris M, Rose S, Baer A, et al. Challenges in adapting existing clinical natural language processing systems to multiple, diverse health care settings. J Am Med Inform Assoc. 2017 Sep 1;24(5):986–91. doi: 10.1093/jamia/ocx039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson SB, Adekkanattu P, Campion Jr. TR, Flory J, Pathak J, Patterson OV, et al. 2017. Nov, From Sour Grapes to Low-Hanging Fruit: A Case Study Demonstrating a Practical Strategy for Natural Language Processing Portability. AMIA Annu Symp Proc. [PMC free article] [PubMed] [Google Scholar]
- 19.Divita G, Carter ME, Tran L-T, Redd D, Zeng QT, Duvall S, et al. v3NLP Framework: Tools to Build Applications for Extracting Concepts from Clinical Text. EGEMS (Wash DC) 2016 Aug 11;4(3):1228. doi: 10.13063/2327-9214.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.HEDIS 2016 Final NDC Lists [Internet] [cited 2018 Mar 2]. Available from: http://www.ncqa.org/hedis-quality-measurement/hedis-measures/hedis-2016/hedis-2016-ndc-license/hedis-2016-final-ndc-lists.
- 21.Patterson OV, Freiberg MS, Skanderson M J, Fodeh S, Brandt CA, DuVall SL. Unlocking echocardiogram measurements for heart disease research through natural language processing. BMC Cardiovasc Disord. 2017 Jun 12;17(1):151. doi: 10.1186/s12872-017-0580-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garvin JH, DuVall SL, South BR, Bray BE, Bolton D, Heavirland J, et al. Automated extraction of ejection fraction for quality measurement using regular expressions in Unstructured Information Management Architecture (UIMA) for heart failure. J Am Med Inform Assoc. 2012 Oct;19(5):859–66. doi: 10.1136/amiajnl-2011-000535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sholle ET, Kabariti J, Johnson SB, Leonard JP, Pathak J, Varughese VI, et al. 2017. Secondary use of patients’ electronic records (SUPER): an approach for meeting specific data needs of clinical and translational researchers. AMIA Annu Symp Proc. [PMC free article] [PubMed] [Google Scholar]
- 24.Bowman S. Impact of electronic health record systems on information integrity: quality and safety implications. Perspect Health Inf Manag. 2013 Oct 1;10:1c. [PMC free article] [PubMed] [Google Scholar]
- 25.Mo H, Thompson WK, Rasmussen LV, Pacheco JA, Jiang G, Kiefer R, et al. Desiderata for computable representations of electronic health records-driven phenotype algorithms. J Am Med Inform Assoc. 2015 Nov;22(6):1220–30. doi: 10.1093/jamia/ocv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richesson RL, Smerek MM, Blake Cameron C. A framework to support the sharing and reuse of computable phenotype definitions across health care delivery and clinical research applications. EGEMS (Wash DC) 2016 Jul 5;4(3):1232. doi: 10.13063/2327-9214.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conway M, Berg RL, Carrell D, Denny JC, Kho AN, Kullo IJ, et al. Analyzing the heterogeneity and complexity of Electronic Health Record oriented phenotyping algorithms. AMIA Annu Symp Proc. 2011 Oct 22;2011:274–83. [PMC free article] [PubMed] [Google Scholar]