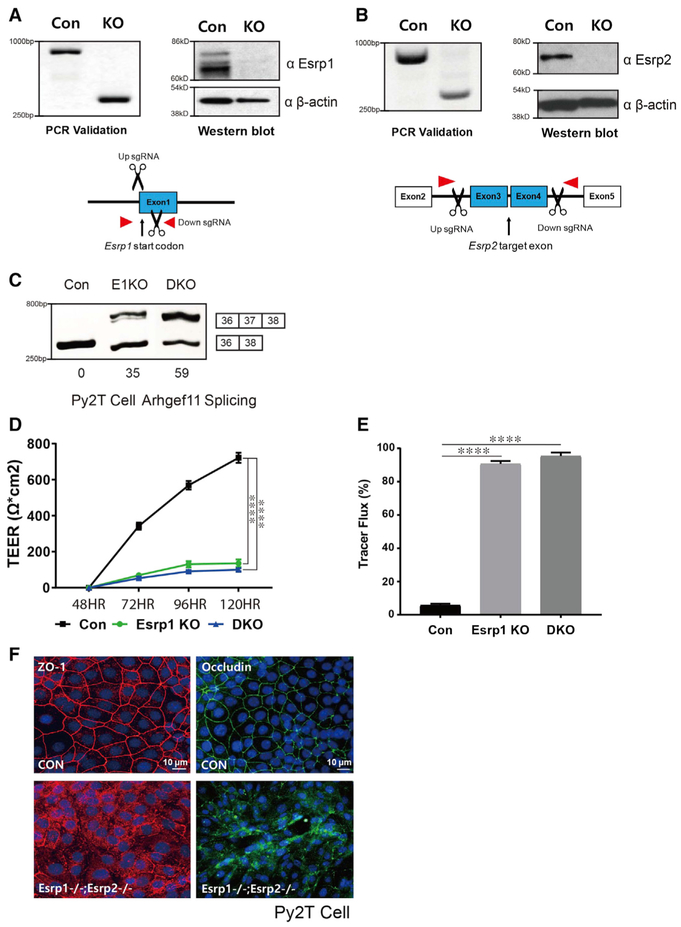
Figure 4. Generation of CRISPR/Cas9-Mediated Esrp KO in Epithelial Cells.
(A and B) Schematics and validation using CRISPR/Cas9 to ablate Esrp1 (A) and Esrp2 (B) in Py2T cells at the level of genomic DNA and protein by western blot. For Esrp1, a Cas9 nickase strategy was used to target the Esrp1 start codon region, whereas standard Cas9 was used to delete exons in Esrp2 that would disrupt the reading frame.
(C) RT-PCR analysis of Arhgef11 exon 37 splicing in control, Esrp1−/− knockout (KO), Esrp1−/−; Esrp2−/− DKO Py2T cells. Values for exon 37 percentage spliced in (PSI) are indicated beneath each lane.
(D) TEER was measured across confluent control or KO epithelial cell monolayers that had been stimulated with calcium for the indicated times.
(E) FITC-dextran tracer flux of control, Esrp1 KO, or Esrp1/2 DKO epithelial cells. Error bars indicate means ± SDs, n = 3. Statistical significance comparing each group with control was determined by t test. ****p < 0.0001.
(F) Disrupted Occludin localization in Esrp1−/−;Esrp2−/− DKO Py2T epithelial cells after Ca2+ switch.