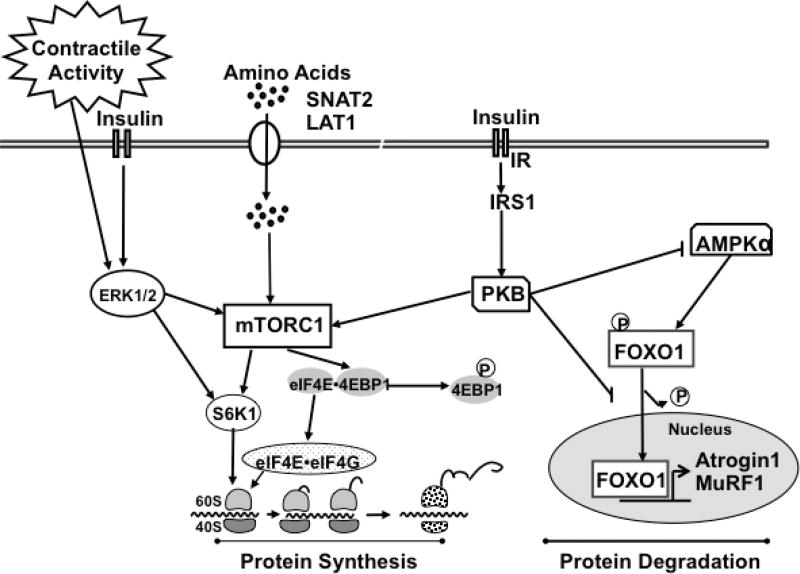
Fig. 1.
Schematic representation of protein synthesis and degradation pathways in skeletal muscle. Briefly, insulin signals by activating the insulin receptor (IR) leading to protein kinase B activation (PKB). Upon activation, mTORC1 phosphorylates its downstream effectors, ribosomal protein S6 kinase 1 (S6K1) and eIF4E-binding protein-1 (4EBP1). Phosphorylated 4E-BP1 releases eIF4E from the inactive eIF4E·4E-BP1 complex to form the active eIF4G·eIF4E complex that binds to mRNA and initiates translation. Amino acid signaling toward protein synthesis is less well understood; nonetheless, the consensus is that amino acids activate substrates downstream of mTOR, leading also to an increase in protein translation initiation via S6K1 and eIF4G· eIF4E activation. Extracellular signal-regulated kinases 1 and 2 (ERK1/2) also activate the mTOR pathway in response to insulin and contractile activity. In addition, insulin inactivates the transcription factors of the forkhead box O (FOXO) family and AMP-activated protein kinase α (AMPKα), which in turn downregulates the transcription of the E3 ubiquitin ligases muscle Atrophy F-Box/Atrogin-1 (MAFbx/Atrogin-1) and muscle RING-finger protein-1 (MuRF1), decreasing overall protein degradation.