FIGURE 3.
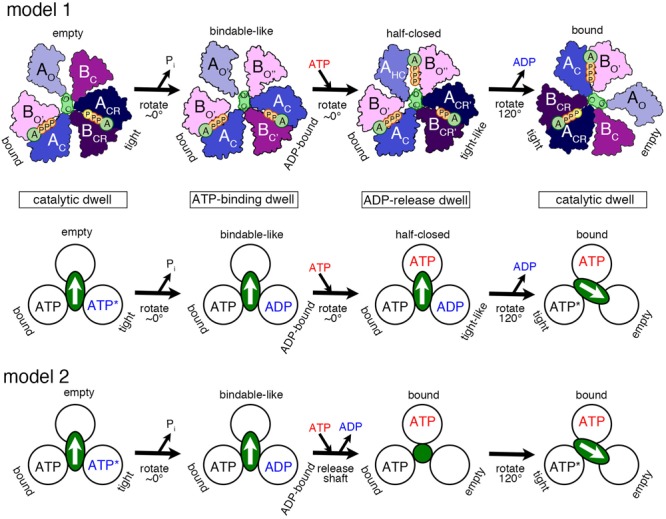
Proposed models of the rotation mechanism of Enterococcus hirae V1-ATPase based on the crystal structures. [Model 1 (Suzuki et al., 2016)] Upper drawings show the structural models from the left to the right panel based on the crystal structures of 2ATPV1 (catalytic dwell), 2ADPV1 (ATP-binding dwell), 3ADPV1 (ADP-release dwell), and 2ATPV1. ATP indicated with a yellow ‘P’ in 2ATPV1 represents an ATP molecule that is committed to hydrolysis. Lower drawing shows the coupling model of the 120° rotation of the shaft (green ellipse with white arrow) and the ATP hydrolysis based on the structural model (upper drawings). Each circle represents the conformation of the nucleotide-binding site, viewed from the cytoplasmic side. The orientation of the shaft begins from the 12 o’clock position in the catalytic dwell waiting for ATP hydrolysis. ATP∗ represents an ATP molecule that is committed to hydrolysis. ATP∗ is hydrolyzed to produce ADP and Pi, and the Pi release induces the conformational changes to the ATP-binding dwell state, without a rotational sub-step. ATP binding at the ‘bindable-like’ form in the ATP-binding dwell state, induces the conformational changes to the ADP-release dwell, without an apparent rotational sub-step. ADP release from the ‘tight-like’ form induces the dissociation of the shaft, thermal 120° rotation, and consequent conformational changes to the catalytic dwell. [Model 2] An alternative coupling scheme of the model 1 without the ADP-release dwell state is shown. ATP binding to the ATP-binding dwell state induces the concomitant release of the shaft and ADP. Therefore, this transient intermediate structure may correspond to that of 2ATPA3B3 with the shaft (green circle) thermally fluctuating. For details, see text.
