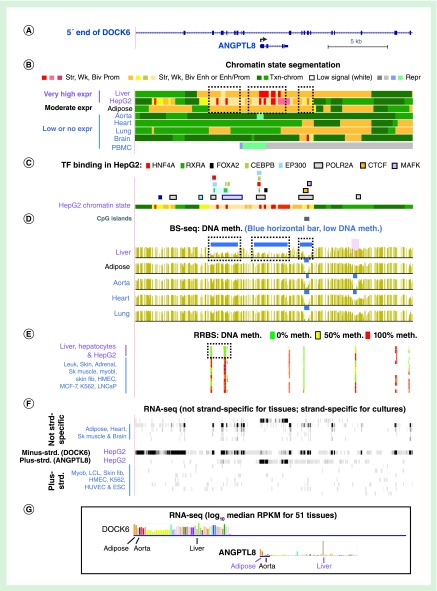
Figure 3. . ANGPTL8 exhibits intragenic enhancer chromatin overlapping DNA hypomethylation specifically in liver, which has the highest expression.
(A) ANGPTL8 is embedded in intron 14 of 47 introns of DOCK6 (chr19:11,339,832–11,363,081). (B–D) Predicted chromatin states, TF binding sites and DNA methylation profile from bisulfite-seq as in Figure 2. Dotted boxes in (B) and (C) indicate regions where liver-associated DNA hypomethylation was seen. (E) DNA methylation profiling by RRBS (with an 11-color gradient) including cell cultures for which bisulfite-seq DNA are not available. (F) and (G), RNA seq data as in Figure 2. MCF-7 and LNCaP, breast and prostate cancer cell lines, respectively; blue highlighting in panel (C), two neighboring RXRA sites and a HNF4A site that overlaps one of them.
ESC: Embryonic stem cell; Expr: Expression; fib: Fibroblast; HepG2: Hepatocarcinoma cell; HMEC: Human mammary epithelial cell; HUVEC: Human umbilical vein endothelial cell; K562: Myelogenous leukemia cells; LCL: Lymphoblastoid cell line; Myob: Myoblast; PBMC: Peripheral blood mononuclear cells; RRBS: Reduced representation bisulfite sequencing; Sk muscle: Skeletal muscle; TF: Transcription factor.