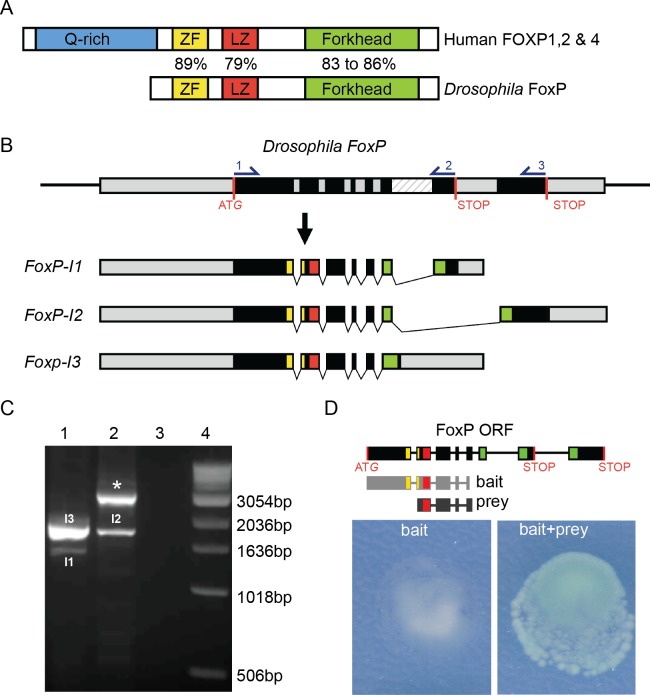
Fig 1. Conserved properties of Drosophila FoxP.
(A) Schematic representation of human FOXP and Drosophila FoxP proteins. FOXP protein domains: glutamate-rich region (Q-rich, in blue), zinc finger (ZF, in yellow), leucine zipper (LZ, in red) and forkhead domain (Forkhead, in green) are indicated. % indicates aa similarity between human FOXP1, 2, 4 and Drosophila FoxP protein domains. (B) The Drosophila FoxP genomic region (exons in black, untranslated regions (UTRs) and introns in grey, and intron 6 who’s retention gives rise to FoxP-I3 in a striped pattern, START and alternative STOP codons in red) and the three encoded transcripts (FoxP-I1 to–I3). Protein domain-encoding regions are highlighted using the color code used in (A). Primers for RT-PCR analysis are indicated with numbers in the FoxP genomic region. (C) Agarose gel analysis of RT-PCR products amplified with primers 1–2 in lane 1 (PCR products corresponding to FoxP-I1 (I1, 1329bp) and -I3 (I3, 1701bp)) and primers 1–3 in lane 2 (lower band corresponds to FoxP-I2 (I2, 2999bp), upper band (*) corresponds to an amplicon derived either from an unspliced FoxP pre-mRNA or amplification of genomic DNA present in the sample (2824bp)). Lane 3: negative control (primers, no template). Lane 4: molecular weight marker. (D) FoxP-FoxP dimerization in the yeast two-hybrid assay. The utilized construct (light grey) and isolated FoxP fragment (prey, dark grey) are depicted. The yeast two-hybrid bait alone shows no autoactivation and growth. When yeast are co-transformed with both, bait and prey induce colony growth and β-galactosidase activity, demonstrating FoxP dimerization.