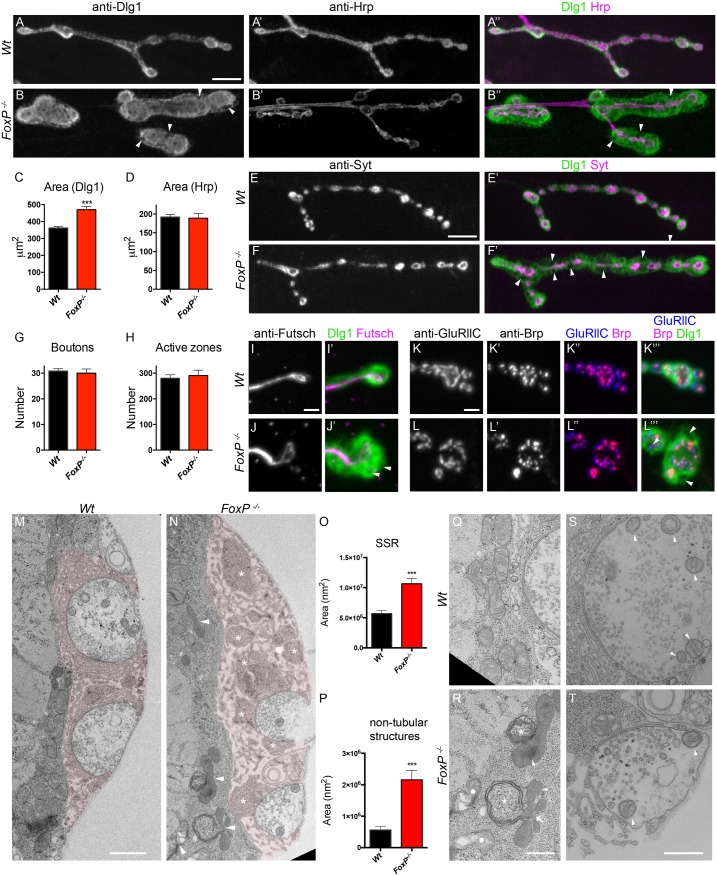
Fig 6. FoxP regulates NMJ postsynaptic morphology.
Muscle four type 1b NMJs of FoxP-/- mutant and wildtype wandering L3 male larvae. (A-B) Co-immunostaining of Dlg1 and Hrp. Scale bar: 10μm. Dlg1 staining showing a honeycomb-like pattern, disorganization and covering a wider region at FoxP-/- mutant synaptic terminals compared to wildtype (Wt) terminals. (C) Dlg1 synaptic area is significantly increased in FoxP-/- mutants (wt n = 56, FoxP-/- n = 60). (D) Hrp-labelled synaptic area does not differ between FoxP-/- and wildtype (wt n = 27, FoxP-/- n = 23). (E-F) Co-immunostainings of Syt and Dlg1. Scale bar: 10μm. (G) The number of synaptic boutons (wt n = 55, FoxP-/- n = 35) and (H) the number of active zones (wt n = 18, FoxP-/- n = 19) do not differ between FoxP-/- and wildtype. (I, J) Co-immunostainings of Futsch and Dlg1. Scale bar: 2μm and (K, L) Co-immunostainings of Brp, GluRllC and Dlg1. Scale bar: 2μm. Syt, Futsch, Brp and GluRllC NMJ immunolabeling appear normal in FoxP-/-. Electron micrographs of third instar larvae NMJ synaptic boutons (muscle 6/7), (M) wildtype and (N) FoxP mutants. SSR surrounding synaptic boutons was shaded in pale red using Adobe illustrator. Asterisks indicate non-tubular structures located in the SSR, arrowheads indicate defective mitochondria. Scale bar: 1 μm. (O) SSR area is significantly increased in FoxP-/- (wt n = 39, FoxP-/- n = 30). (P) The area occupied by non-tubular structures is significantly increased in FoxP-/- (wt n = 40, FoxP-/- n = 30). (Q-R) Mitochondria surrounding the SSR present ultrastructural defects in FoxP-/- mutants. Arrowheads indicate defective cristae; arrows indicate multilobar mitochondria; asterisks indicate membranes folds around the mitochondria resembling autophagosomal structures, circles indicate collapsed mitochondria. Scale bar: 250 nm. (S-T) The conformation of neuronal mitochondria is unaffected in the FoxP-/-. Arrowheads indicate mitochondria. Scale bar: 500 nm. Bars represent the mean, error bars indicate SEM, t-test between conditions was performed for each parameter to determine significance (***, p<0.001). For the underlying numerical data see S9 and S10 Tables.