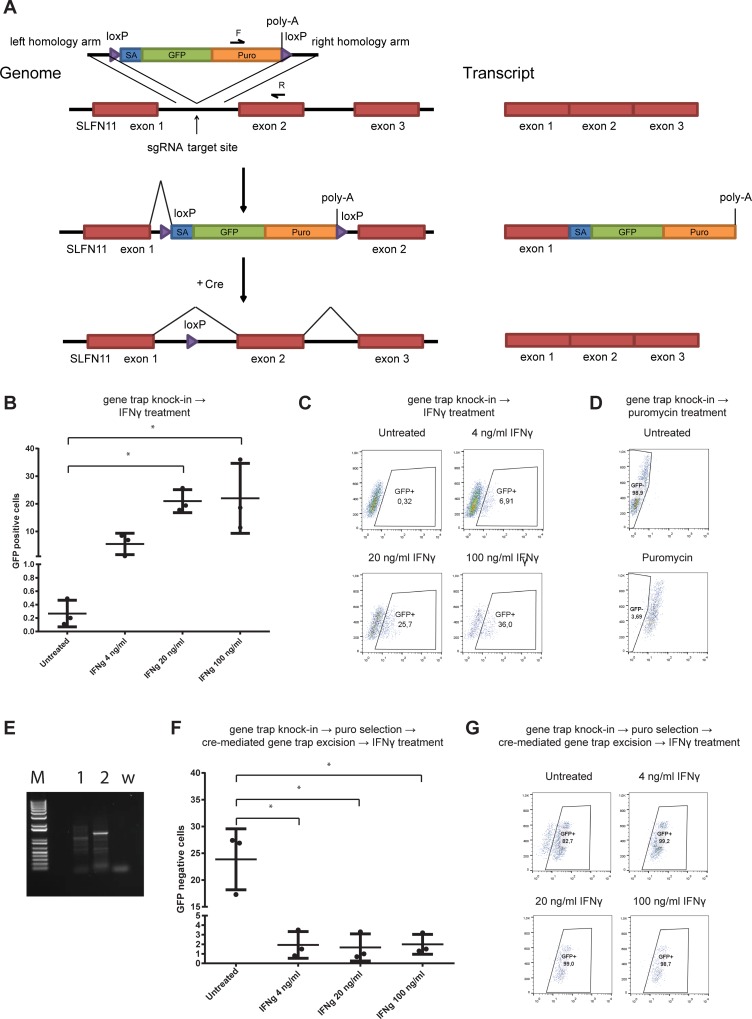
Fig 7. Reversible inhibition of IFN-γ sensitivity by inactivation of the SLFN11 locus.
A) Schematic overview of experimental design. A promoterless gene trap vector was inserted into the first intron of the SLFN11 gene. SLFN11 exons are indicated in red, GFP and puromycin N-acetyltransferase coding sequences in green and orange, and loxP sites in purple. Left part of the panel depicts the genomic locus, right part depicts the resulting transcripts. B-C) Cells with integrated gene trap vector were either left untreated or exposed to the indicated IFN-γ concentrations. Technical replicates (B) and representative flow plots (C) are depicted. D) Puromycin treatment results in homogeneous selection of cells with integrated gene trap vector. E) Detection of the integrated gene trap vector in the SLFN11 locus by PCR. Used primers are depicted in panel A (F and R). 1: untransfected HAP1; 2: transfected and puromycin-selected HAP1; w: water control. F-G) Excision of the gene trap vector results in re-sensitization of HAP1 cells to IFN-γ. Cells from panel D were transfected with a cre expressing vector and either left untreated or exposed to the indicated IFN-γ concentrations. Technical replicates (E) and representative flow plots (F) are depicted. * p<0.05.