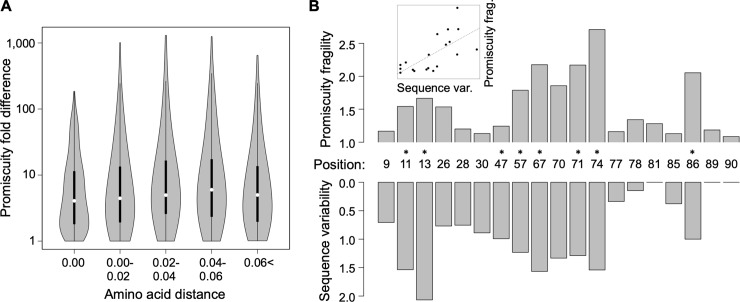
Fig 3. Promiscuity changes rapidly during evolution and might be a selectable trait.
(A) For all pairs of selected alleles, the predicted promiscuity difference between two HLA-DRB1 alleles is shown as a function of amino acid distance measured after excluding the epitope-binding region. Large differences in promiscuity can be observed even between closely related pairs of alleles (e.g., at zero amino acid distance). As a result, there is no correlation between amino acid distance and promiscuity fold difference (Spearman’s rho = 0.02, P = 0.19). Amino acid distances were binned as shown on the figure (n = 308, 1,168, 564, 654, 1,492). Violin plots show the density function of promiscuity fold difference values for allele pairs in the given bin. White circles show median values; bold black lines show the interquartile range. (B) Sequence variability of an amino acid site in the epitope-binding region of HLA-DRB1 (measured as Shannon entropy) correlates positively with the site’s promiscuity fragility, measured as the median predicted promiscuity fold difference caused by a random amino acid change at the given site (see inset, Spearman’s rho: 0.76, P = 0.0001). Sites that have a larger impact on promiscuity are more diverse in human populations. Line in inset represents linear regression between the two variables. The same result was obtained when promiscuity fragility was calculated based on nucleotide substitutions instead of amino acid substitutions (Spearman’s rho: 0.73, P = 0.0004, see Methods) or when sequence variability was measured as nonsynonymous nucleotide diversity (πA) instead of sequence entropy (S7 Fig). Sites under positive selection as identified by Furlong and colleagues [48] show significantly higher promiscuity fragility (Wilcoxon rank sum test, P = 0.0012) and are marked with asterisks (see also S8 Fig). The underlying data for this figure can be found in S4 Data.