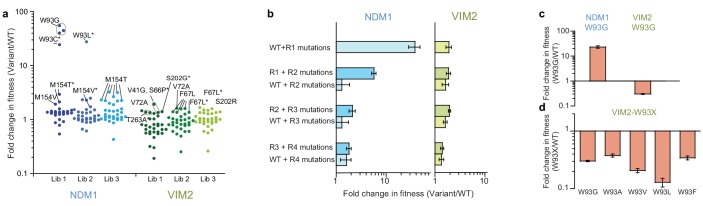
Figure 2. Accessibility of adaptive mutations from each starting point.
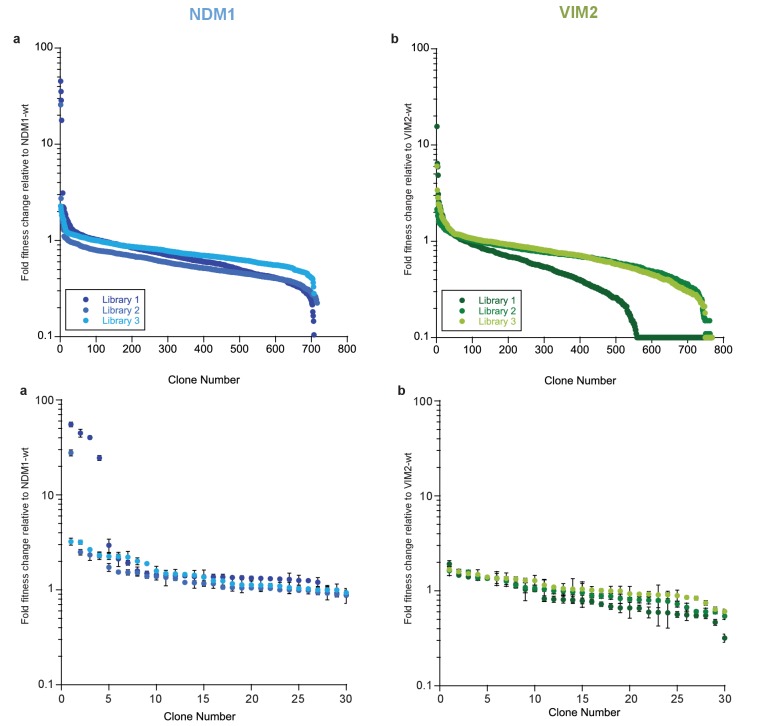
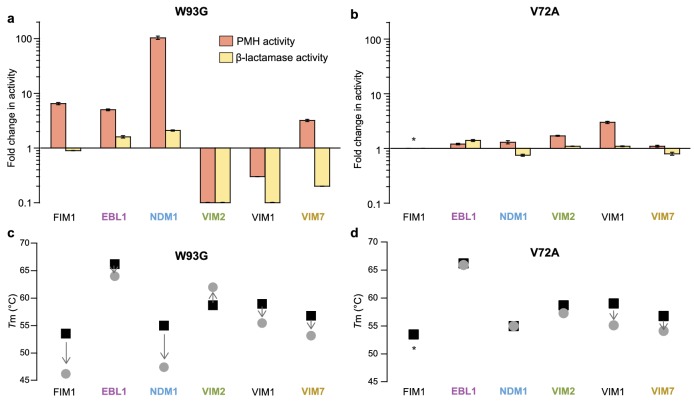
(A) Additional screenings of NDM1 and VIM2 round one libraries. Activities of the top 30 variants isolated from additional screenings of three independently generated mutagenized libraries and normalized on the PMH fitness of NDM1-WT and VIM2-WT, respectively; presented as scatterplots. Mutants are labeled when variants possess a mutation previously encountered in the original directed evolution or if this mutation is located at the same position. Asterisks indicate that the variant has other mutations in addition to the labeled one. Mutations and activities of all improved variants are listed in Supplementary file 1H. The activities of all 2160 variants in the round one additional screenings are presented in Figure 2—figure supplement 1. (B) Epistasis analysis of mutations. Mutations occurring in the NDM1 and VIM2 trajectories were introduced into their respective wild-type sequence and the resulting PMH fitness was compared to the original PMH fitness change observed during the trajectory. (C) Fold change in PMH fitness induced by mutation W93G in NDM1-WT and VIM2-WT, respectively. (D) Fold change in PMH fitness induced by mutation of Trp93 in VIM2 to various hydrophobic residues. A full description of the lysate activities is presented in Supplementary file 1G. Errors bars represent the propagated standard deviation from triplicate measurements.