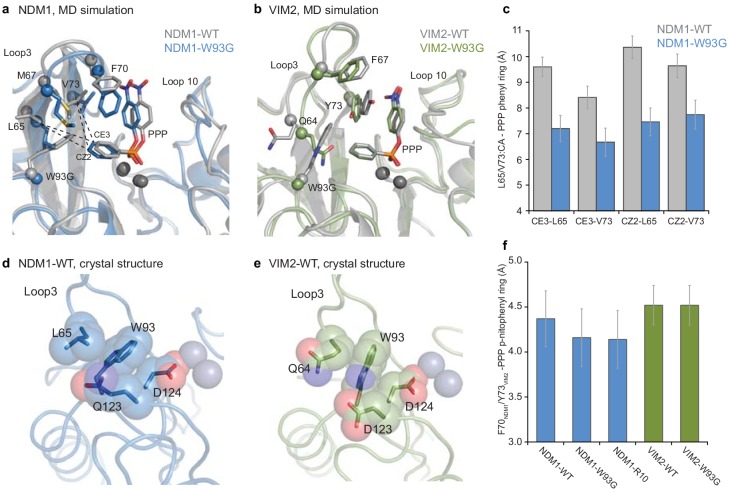
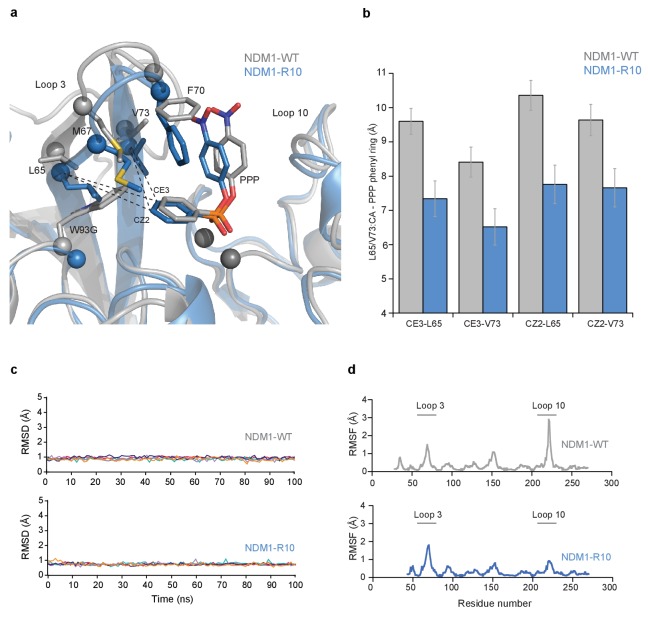
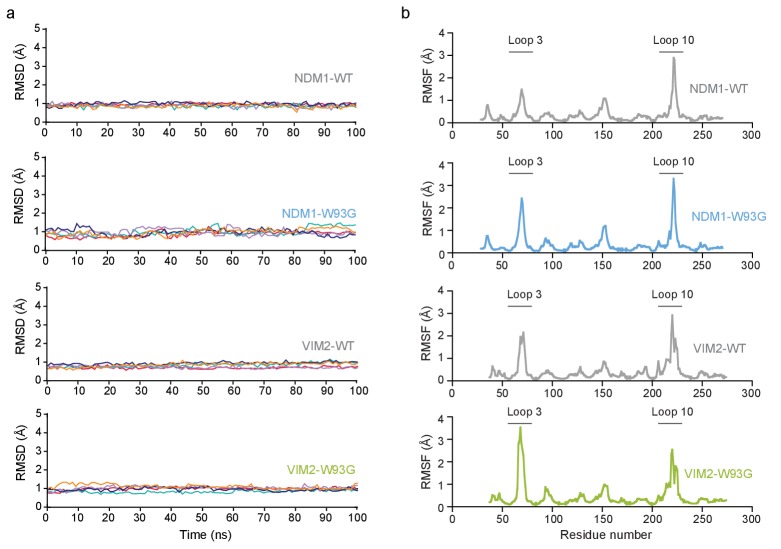
Figure 5. Comparisons of the structural changes induced by mutation W93G.
Representative structures from MD simulations of (A) NDM1-WT (grey) and NDM1-W93G (blue), and (B) VIM2-WT (grey) and VIM2-W93G (green). (C) Changes in hydrophobic interactions between the phenyl ring of the PMH substrate (CE3 and CZ2 atoms) and residue Leu65 and Val73 (α-carbon) of NDM1 upon W93G, during the MD simulations. (D and E) Comparison of the position of Trp93 and second shell residues in the (D) NDM1-WT (PDB-ID: 3SPU) and (E) VIM2-WT (PDB-ID: 1KO3) crystal structures. (F) Average distance between the p-nitrophenyl ring of the PMH substrate and the side chain rings of Phe70 in NDM1, and Tyr73 in VIM2, when the two rings form a π-π interaction during the MD simulations.