Fig. 2.
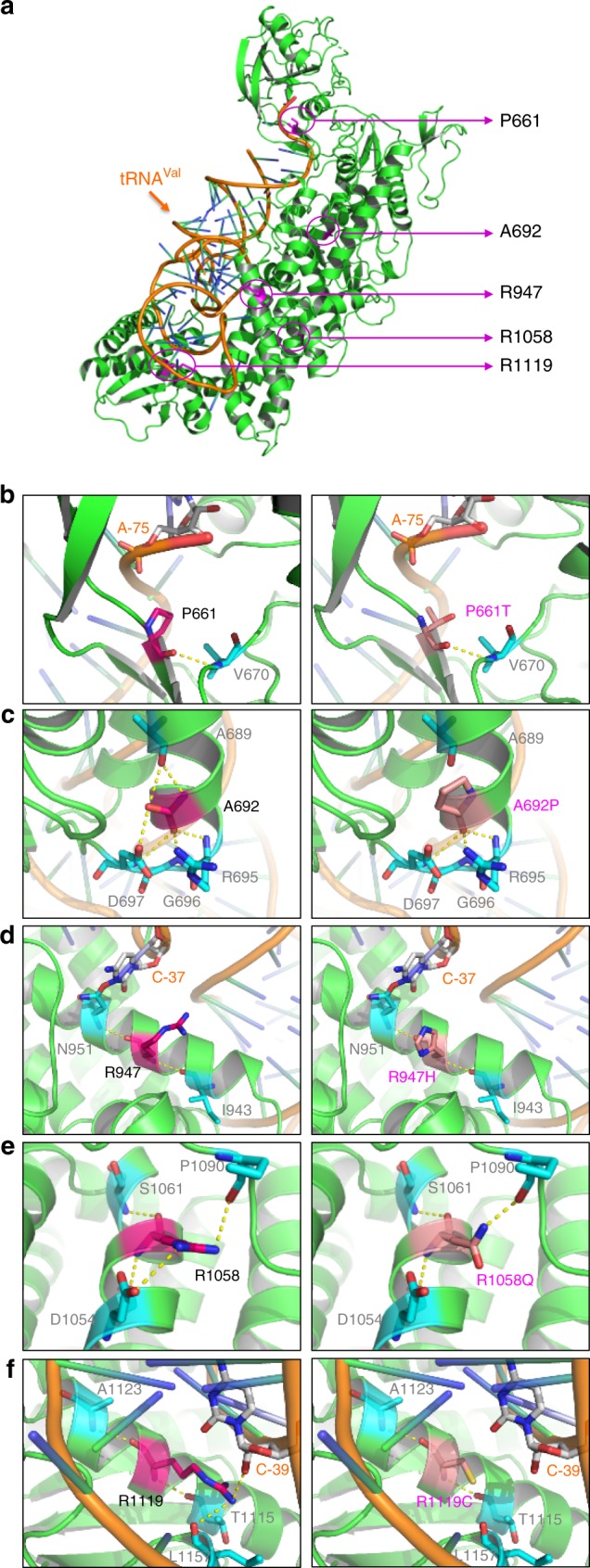
Predicted mutational impact on VARS structure and tRNA binding. a Locations of the mutational sites on the structural model of human VARS-tRNAVal complex (VARS green, tRNAVal orange, mutational sites magenta). Residues Pro661, Arg947, and Arg1119 are closer to the tRNA molecule (<6 Å), while Ala692 and Arg1058 have a longer distance of >15 Å from tRNA. b–f Pair-wise comparisons between the wild-type (left) and mutant (right) residues for predicted changes in local contacts with tRNA or other amino acids. Amino acids are indicated by letter and number (e.g., Pro661), tRNA bases are indicated by letter and dash and number (e.g., A-75). Hydrogen bonds are presented as yellow dashed lines. b Residue Pro661 forms a hydrogen bond with Val670 and has a distance of ~4.5 Å from A-75 of the tRNA. c Residue Ala692 forms hydrogen bonds with adjacent residues Ala689, Arg695, Gln696, and Asp697. Mutation to proline predicts abolished contacts with Ala689 and disrupted helical structure. d Residue Arg947 is close to C-37 and predicts hydrogen-bonding interaction that is abolished by the Arg947H mutation. This mutation does not predict altered hydrogen-bonding with Ile943 and Asn951. e Residue Arg1058 forms hydrogen bonds with Asp1054, Ser1061 and Pro1090. The NM_006295.2:c.3173G>A p.(Arg1058Gln) mutation predicts elimination one of the two possible bonds with Asp1054. f The side chain of Arg1119 forms a hydrogen bond with C-39, predicted to be abolished by the mutation
