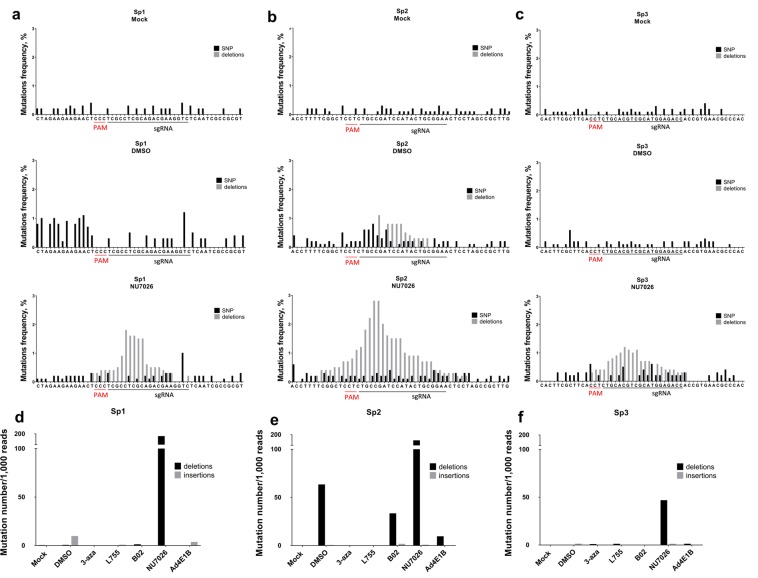
Figure 4.
Deep sequencing of CRISPR/Cas9 on-target sites. (a–c) Mutation rates and profiles of DSB repair outcomes in the target regions of HBV cccDNA isolated from HepG2-1.1merHBV cells. Each sgRNA (Sp1, Sp2, Sp3) has a mock control group, DMSO control group, and groups treated with NU7026 (see Supplementary Fig. S4 for results of treatment with other small molecules). Mock group was transfected with a Cas9 vector and non-targeting PCR product and treated with DMSO. Control groups were transfected with a Cas9 vector and an HBV-targeting sgRNA encoded by a PCR product, and treated with DMSO. PAM sequence for all sgRNAs was NGG (the strand complementary to the target DNA is provided). Regions containing the on-target sites were sequenced, and frequencies of insertions/deletions were calculated for sgRNAs Sp1 (d), Sp2 (e), and Sp3 (f) in cells treated with 3-aza, L755, B02, and Ad4E1B compared to mock control group and to DMSO control group. Number of indels per 1000 reads was counted for all experimental groups with corresponding sgRNAs.